The new Data Panel goes beyond a simple platform
for analyzing and visualizing input data. Besides
allowing the analysis and visualization of input variables, it now also
allows the visualization and analysis of all your models and
derived variables,
giving you access to a rich web of analyses between all these types of variables and
across different datasets.
The myriad of analyses supported by the new Data Panel includes scatter plots and
regression analysis
between all possible pairs of variables (input variables, models and derived variables);
histograms and summary statistics for all variables and across different datasets;
different line charts for outlier detection and model analysis;
evaluation of the variable importance and
display of the
variable importance chart for all the models; and so on.
The new Data Panel also supports extensive record analyses, allowing you to study
different types of records using different charts and browsing tools. For example,
you can now compare each record with different record prototypes in order to gain insight
into both your data and your models.
By selecting just the outliers or the
misclassified records to browse, you can now also do
error analysis in the Data Panel.
Variable Charts and Analysis
Below is a gallery of the new charts of the Data Panel for variable visualization
& analysis.
The Sequential Distribution Chart, with its standard deviation lines and average line,
is ideal for detecting outliers. The Bivariate Line Chart, with its sorting options
and Show Normalized option, is a
very flexible and useful tool for comparing any two variables.
The Scatter Plot
allows you to very quickly visualize the correlation between all possible pairs
of variables, including input variables, models and derived variables.
The Histogram allows you to very quickly analyze the distribution of all your variables
(input variables, models and derived variables),
and compare them across different datasets.
The Statistics Charts, which include 9 different charts for 9 different summary statistics
(min, max, average, median, standard deviation, R-square, correlation coefficient,
slope and intercept), allow you to quickly visualize and compare summary statistics for
all your variables (input variables, models and derived variables) and for all your datasets.
The Variable Importance Chart shows the importance of all the variables in your models,
including the importance of derived variables.
The new highlighting functionality of the new Data Panel offers another dimension to
data visualization and analysis. By highlighting certain types of records,
such as misclassifications in Classification and Logistic Regression or
model outliers in
Regression and Time Series Prediction, you can now quickly understand and visualize
new patterns in all your variables and models.
Record Charts and Analysis
Below is a gallery of different record analyses you can
easily perform in the new Data Panel.
By combining all the Record Charts with different browsing options, GeneXproTools now allows
you to perform different record analyses and computations, including error analysis, record prototyping and
summary statistics across different variables (input variables, models and
derived variables) and datasets.
 |
Support for Categorical Variables and Missing Values |
GeneXproTools now supports categorical variables and 50+ different types of missing values,
replacing them automatically during data loading, but allowing you to choose different
mappings both for the categories and missing values.
Moreover GeneXproTools extends the support for categorical and missing data throughout
the modeling process, generating model code that also supports categorical variables and
missing values, offering you a much more robust and convenient platform for model design
and deployment. Below is an example in C++ of a classification model with
both categorical variables and missing values.
//------------------------------------------------------------------------
// Classification model generated by GeneXproTools 5.0 on 6/8/2013
// GEP File: D:\GeneXproTools\Version5.0\CreditApproval_CM_01.gep
// Training Records: 460
// Validation Records: 230
// Fitness Function: Bounded ROC, ROC Threshold
// Training Fitness: 912.951824483014
// Training Accuracy: 87.83% (404)
// Validation Fitness: 950.172009488309
// Validation Accuracy: 90.43% (208)
//------------------------------------------------------------------------
#include "math.h"
int gepModel(double d[]);
double gep3Rt(double x);
double gepMin2(double x, double y);
void TransformCategoricalInputs(char* input[], double output[]);
int gepModel(char* d_string[])
{
const double ROUNDING_THRESHOLD = 2.02223099900018;
const double G1C2 = 0.481887264625996;
const double G1C6 = 3.80779442732017;
double d[15];
TransformCategoricalInputs(d_string, d);
double dblTemp = 0.0;
dblTemp = atan((((d[12]*(d[7]-d[13]))-((exp(G1C6)+d[3])/2.0))-G1C2));
dblTemp += gep3Rt(pow(gepMin2(d[8],d[9]),3));
dblTemp += atan((d[9]*gep3Rt(((pow(d[7],2)-(d[9]-d[14]))
-pow((d[4]+d[4]),4)))));
dblTemp += pow(d[8],2);
return (dblTemp >= ROUNDING_THRESHOLD ? 1 : 0);
}
double gep3Rt(double x)
{
return x < 0.0 ? -pow(-x,(1.0/3.0)) : pow(x,(1.0/3.0));
}
double gepMin2(double x, double y)
{
double varTemp = x;
if (varTemp > y)
varTemp = y;
return varTemp;
}
void TransformCategoricalInputs(char* input[], double output[])
{
if(strcmp("l", input[3]) == 0)
output[3] = 1.0;
else if(strcmp("u", input[3]) == 0)
output[3] = 2.0;
else if(strcmp("y", input[3]) == 0)
output[3] = 3.0;
else if(strcmp("?", input[3]) == 0)
output[3] = 2.0;
else output[3] = 0.0;
if(strcmp("g", input[4]) == 0)
output[4] = 2.0;
else if(strcmp("gg", input[4]) == 0)
output[4] = 1.0;
else if(strcmp("p", input[4]) == 0)
output[4] = 3.0;
else if(strcmp("?", input[4]) == 0)
output[4] = 2.0;
else output[4] = 0.0;
output[7] = atof(input[7]);
if(strcmp("f", input[8]) == 0)
output[8] = 1.0;
else if(strcmp("t", input[8]) == 0)
output[8] = 2.0;
else output[8] = 0.0;
if(strcmp("f", input[9]) == 0)
output[9] = 1.0;
else if(strcmp("t", input[9]) == 0)
output[9] = 2.0;
else output[9] = 0.0;
if(strcmp("g", input[12]) == 0)
output[12] = 1.0;
else if(strcmp("p", input[12]) == 0)
output[12] = 2.0;
else if(strcmp("s", input[12]) == 0)
output[12] = 3.0;
else output[12] = 0.0;
if(strcmp("?", input[13]) == 0)
output[13] = 0.0;
else output[13] = atof(input[13]);
output[14] = atof(input[14]);
}
 |
Support for Multinomial Classification & Logistic Regression |
GeneXproTools now provides a platform for handling multiple classes in Classification
and Logistic Regression, allowing you to easily setup different sub-classification
tasks for each class in the response variable, without you having to prepare and load different
datasets for each Classification or Logistic Regression problem. Through the
Class Merging & Discretization window you can single out the class of interest and
then create models for each sub task, keeping them under a single gep file or creating
n different files for the n classes.
 |
Support for Data Normalization |
GeneXproTools now allows you to normalize your numerical variables, not only
for purposes of visualization and analysis but also for model design. The
normalization techniques supported by GeneXproTools include Standardization,
0/1 Normalization and Min/Max Normalization.
GeneXproTools again creates model code for scoring and deployment that
also supports data normalization. Below is an example in Python of a
logistic regression model created using data standardized in the GeneXproTools environment.
#------------------------------------------------------------------------
# Logistic regression model generated by GeneXproTools 5.0 on 6/9/2013
# GEP File: D:\GeneXproTools\Version5.0\Diabetes-DN_01a.gep
# Training Records: 512
# Validation Records: 256
# Fitness Function: Maximum Likelihood, Logistic Threshold
# Training Fitness: 616.228156623755
# Training Accuracy: 79.69% (408)
# Validation Fitness: 546.661612370381
# Validation Accuracy: 80.08% (205)
#------------------------------------------------------------------------
from math import *
def gepModel(d_string):
ROUNDING_THRESHOLD = -1.77709077258152
G2C8 = 2.57484664448988
G3C0 = -6.54957731864376
G3C9 = 2.25135044404431
G3C7 = 5.69697519760735
G4C2 = 8.97033600878933
G4C7 = 5.79271828363903
G4C4 = 1.07699819940794
d = [0.0] * len(d_string)
TransformCategoricalInputs(d_string, d)
Standardize(d)
varTemp = 0.0
varTemp = (d[1]-(1.0-pow(d[1],3.0)))
varTemp = varTemp + min((((pow(d[5],3.0)+d[1])/2.0)*G2C8),\
(gep3Rt(d[4])-atan(d[2])))
varTemp = varTemp + max(max((gep3Rt(G3C7)+d[6]),(G3C0*G3C9)),\
((G3C0+pow(d[5],3.0))/2.0))
varTemp = varTemp + (((((pow(d[7],2.0)+d[1])/2.0)+(G4C2+G4C7))/2.0)\
*min(d[7],(1.0-G4C4)))
if (varTemp >= ROUNDING_THRESHOLD):
return 1
else:
return 0
def gep3Rt(x):
if (x < 0.0):
return -pow(-x,(1.0/3.0))
else:
return pow(x,(1.0/3.0))
def TransformCategoricalInputs(inputList, outputList):
outputList[1] = {
"?" : lambda : 123.065088757397,
}.get(inputList[1], lambda : float(inputList[1]))()
outputList[2] = {
"?" : lambda : 72.5246913580247,
}.get(inputList[2], lambda : float(inputList[2]))()
outputList[4] = {
"?" : lambda : 156.600790513834,
}.get(inputList[4], lambda : float(inputList[4]))()
outputList[5] = {
"?" : lambda : 32.7155069582505,
}.get(inputList[5], lambda : float(inputList[5]))()
outputList[6] = float(inputList[6])
outputList[7] = float(inputList[7])
def Standardize(input):
AVERAGE_1 = 123.065088757396
STDEV_1 = 30.5208809235629
input[1] = (input[1] - AVERAGE_1) / STDEV_1
AVERAGE_2 = 72.5246913580247
STDEV_2 = 11.6112169688815
input[2] = (input[2] - AVERAGE_2) / STDEV_2
AVERAGE_4 = 156.600790513834
STDEV_4 = 83.1959782804369
input[4] = (input[4] - AVERAGE_4) / STDEV_4
AVERAGE_5 = 32.7155069582505
STDEV_5 = 6.54802505977307
input[5] = (input[5] - AVERAGE_5) / STDEV_5
AVERAGE_6 = 0.470236328125
STDEV_6 = 0.312962638849511
input[6] = (input[6] - AVERAGE_6) / STDEV_6
AVERAGE_7 = 33.478515625
STDEV_7 = 12.0024266685208
input[7] = (input[7] - AVERAGE_7) / STDEV_7
 |
Dataset Partitioning and Sub-sampling |
Now in version 5.0 you can manage your datasets inside GeneXproTools, as it provides essential
tools for you to easily split your data into training and validation/test datasets.
The sampling schemes supported by GeneXproTools include Odds/Evens splitting, choosing
different Partitions and Random Shuffle (random sampling without replacement).
For all modeling categories, GeneXproTools also provides useful defaults optimized for
efficiency and good model generalization.
Moreover GeneXproTools also supports sub-sampling of both the training and
validation datasets, implementing several sub-sampling schemes, including
bagging and mini-batch, which are, respectively, essential tools for creating
random forests (more appropriately called GEP forests) and dealing with large datasets. The sub-sampling of the
validation dataset also has multiple applications, including reserving part
of the validation set for testing and calculating the cross-validation
accuracy of your models.
The sub-sampling of both the training and validation datasets is done in the
Settings Panel and
includes: Top n, Bottom n, Top Half, Bottom Half, Odd Cases, Even Cases, Random, Shuffled,
Balanced Random, and Ballanced Shuffled. The Balanced schemes are available only for classification,
logistic regression and logic synthesis.
 |
Support for GEP Files as Data Source |
We’ve now added support for loading data from gep files,
allowing you to choose either the training or the validation/test datasets or
the original data, which in this context is defined as the union of the training
and validation/test datasets. In either case, it’s the original raw data,
with categorical variables and missing values if they exist, that is loaded into GeneXproTools.
Moreover, Time Series Prediction gep files can also be used as source data
for Regression, Time Series Prediction, Classification and Logistic Regression runs,
allowing you to easily create
all kinds of gep runs using the transformed time series as input variables.
 |
Summary Statistics |
Now GeneXproTools provides summary statistics for all input variables, models and
derived variables. The summary statistics are shown in the Statistics Report
in the Data Panel. Moreover, GeneXproTools also shows the summary statistics
(min, max, average, median, standard deviation, correlation coefficient,
R-square, slope and intercept) graphically in the Statistics Charts.
GeneXproTools also evaluates summary statistics for the records, providing
easy ways of comparing different types of records with record prototypes.
For example, by browsing just the misclassified records these statistics and
visualization tools are essential for error analysis.
 |
Outlier Detection & Removal |
In the data Panel, GeneXproTools now offers different tools for outlier detection,
including the new Scatter Plot and the Sequential Distribution Chart, which now shows
both the average line and standard deviation lines for easy detection of outliers.
Moreover, for the Sequential Distribution Chart, GeneXproTools also allows you to copy the
indexes of all the outliers for the current variable by choosing Copy Outlier IDs (3 Sigma)
in the context menu.
The outlier indexes can then be pasted directly into the Delete Records Window
for the easy removal of all outliers.
 |
Regression Analysis |
GeneXproTools allows you to quickly analyze and visualize the correlation
between
all possible combinations of variables in the Data Panel, including input variables,
models and derived variables. Through the Scatter Plot and
Statistics Report of
the Data Panel you have access to the regression line and
regression equation,
the correlation coefficient and R-square for all pairs of variables.
 |
Error Analysis |
GeneXproTools now allows you to do error analysis in the
Data Panel. By selecting
different records to browse, including misclassified records and outliers,
GeneXproTools allows you to analyze these records using different charts to compare
them with other records and record prototypes.
 |
Variable Importance |
GeneXproTools now computes and shows the importance of all the variables for all
your models. The Variable Importance Chart is available through the
Statistics Charts
in the Data Panel. By selecting Model Variables in the Data combobox,
you can quickly access the variables of each model and their relative importance.
The variable importance is computed for all the models GeneXproTools generates,
including regression models, classification and logistic regression models,
and time series prediction models.
 |
Residual Analysis |
GeneXproTools now performs residual analysis for all regression models,
including time series prediction models. The Residuals Plot is accessible
both in the Run Panel and Results Panel.
 |
New Logistic Regression Category |
The Logistic Regression Framework is now implemented as an independent category,
with its own set of fitness functions, visualization tools, and code generation.
Fitness Functions
A total of 60 new
fitness functions for Logistic Regression, including
ROC Measure, Rank Measure, Maximum Likelihood, Hinge Loss, Positive Correl,
R-square, Dual Margin and F Measure, just to name a few.
Most of these fitness functions are multi-objective and include different
adjustable parameters such as the cost matrix, the number of bins, reference simple models,
lower and upper bounds for the model output, parsimony pressure and variable pressure.
New Charts & Statistics
Now in the Run Panel, for up to 1000 records of the training data, you have access to
6 new charts for model design and visualization. These new charts include
the Classification Tapestry, three Binomial Fit Charts with options for showing both
the rounding threshold and misclassifications, the ROC Curve,
and the sophisticated Classification Scatter Plot which is another Gepsoft’s
beautiful innovation. In the gallery below, the same logistic regression model is being analyzed
using the 6 model fitting charts of the Run Panel.
In the Results Panel GeneXproTools now shows the same model fitting charts it shows
in the Run Panel, but now for all the records in the training/validation datasets.
Moreover, for the Binomial Fit Charts, GeneXproTools allows you to plot the
probability[1] in addition to the raw model output.
In the Table of the Results Panel now GeneXproTools shows the Probability[1] and
the Most Likely Class in addition to the Raw Model Output.
Also important is the new set of statistics evaluated in the
Results Panel,
which now include the Classification Accuracy & Error, the Sensitivity & Specificity,
the Positive Predictive Value & Negative Predictive Value, Recall & Precision,
Correlation Coefficient & R-square, Jaccard Similarity, Matthews Correlation Coefficient,
F1 Measure, and the Area Under the ROC Curve.
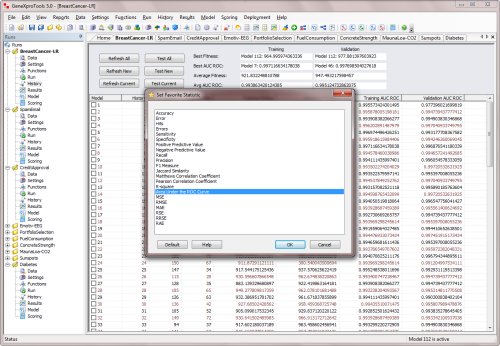
This same set of statistics is available in the History Panel for model and
ensemble selection through the Favorite Statistic Window.
Code Generation
Now GeneXproTools generates the complete code for the logistic regression model.
Moreover, GeneXproTools now lets you choose the kind of output you are most
interested in: the model code for predicting the Probability[1],
the Most Likely Class or the Raw Model Output.
This new modality for code generation is available during model scoring in the Scoring Panel
and also during Model & Ensemble Deployment to Excel.
Also important is that in the Data Panel, for purposes of model visualization and analysis,
you can also choose any of these 3 forms of model output: the Probability[1], the
Most Likely Class or the Raw Model Output.
 |
Model Browsing in the Run Panel |
We’ve now added model navigation in the Run Panel, so now you can go back and forth
analyzing and visualizing your models, not just the model output through the use
of different charts, but also information about the model structure and composition,
such as model size and used variables.
 |
New Charts for Model Visualization & Selection |
We’ve now added 100+ new charts for model visualization & analysis and
improved most of the old ones for all modeling categories. We've added new charts to the Run Panel,
Results Panel and Data Panel of all modeling categories, namely Classification & Logistic Regression,
Regression & Time Series Prediction and Logic Synthesis. Below are highlighted the most important.
Classification & Logistic Regression
In the Run Panel and Results Panel for both Classification & Logistic Regression,
you now have access to six different model fitting charts instead of just the
Classification Tapestry, showing you different aspects of the evolving models,
from the raw model output to the predicted class. You can now analyze and
clearly see the distribution of model outputs relative to the
rounding thresholds using three different Binomial Fit Charts and the new
Classification Scatter Plot. The ROC Curve offers another widely used and
useful dimension to the modeling process and the Classification Tapestry,
which is new for Logistic Regression,
offers a crisp visualization of the model output across the distribution of all
possible outcomes (true positives, true negatives, false positives and false negatives).
In the Data Panel you now have access to additional tools for model selection and analysis,
including the Variable Importance Chart and a total of
nine different Statistics Charts
(min, max, average, median, standard deviation, R-square, correlation coefficient,
slope and intercept) in addition to
all the other new charts of
the new Data Panel for variable and record analysis, including Histograms, Scatter Plots and
different Line Charts. By choosing History Models or
Model Variables or All in the Data combobox in the Data Panel, you can now
perform a myriad of analyses across different datasets and combinations of
models (and model outputs) and variables.
Regression & Time Series Prediction
In the Run Panel and Results Panel we’ve added 3 new charts for
model visualization and
analysis: the Scatter Plot, the Residuals Plot, and the Stacked Distributions Chart.
The Scatter Plot, with the regression line and regression equation, offers a clear view
of the model fit. The Residuals Plot shows the standard residual analysis for
detecting unusual patterns in the distribution of the residuals. The
Stacked Distributions Chart
shows clearly the spread and overlap of the actual and predicted values.
These 3 new charts were also added to the Results Panel, allowing you to perform
the same kind of model analysis for the training and validation/test datasets
and also for different sub-samplings of the training and
validation/test datasets.
In the Data Panel you now have access to additional tools for
model selection and analysis,
including the Variable Importance Chart and a total of
nine different Statistics Charts
(min, max, average, median, standard deviation, R-square, correlation coefficient,
slope and intercept) in addition to
all the other new charts of
the new Data Panel for variable and
record analysis, including Histograms, Scatter Plots and
different Line Charts. By choosing History Models or
Model Variables or All in the Data combobox in the Data Panel, you can now
perform a myriad of analyses across different datasets and combinations of
models and variables.
 |
New Tools for Model Selection |
GeneXproTools now features new tools to help you select your models from basically
any panel, including:
- Access to different Favorite Statistics for all categories;
- Random sub-sampling that enables the application of bootstrap techniques to evaluate with
more confidence the accuracy of your models or any
other measure of fit;
- Model navigation from all panels where models are being analyzed or visualized;
- A new decentralized Delete Active Model functionality that allows you
to delete any model you find lacking in some sense or another;
- The new Rename All Models in the History Panel that allows you
to reindex your models after sorting them using
different statistics, so that you can analyze them
in a particular order, allowing you to gain insights
into their structure and performance;
- New powerful tools for model selection in Time Series Prediction, namely going from
Testing Mode to Prediction Mode and back and changing the number of Testing Predictions,
allowing you to select your models by their performance in the testing data and then use the selected models to make predictions.
 |
Introduction of Evolvable Rounding Thresholds in Classification |
Now GeneXproTools implements the revolutionary idea pioneered by
Candida Ferreira
of different types of evolvable rounding thresholds for classification models.
Now by combining different types of rounding thresholds with different
fitness functions, you have access to a much richer solution space,
allowing you to create even better classifiers. The new evolvable
rounding thresholds GeneXproTools supports include:
 |
Improved Results Panel in all Categories |
We’ve improved the Results Panel in all categories, with more and better charts,
a more extensive set of statistics, and a richer and more interactive table with
different sorting and copying options. For example, the Results Panel for
Classification and Logistic Regression now features 17
Measures of Fit, including
the usual Classification Accuracy and Error, Correlation Coefficient and R-square,
but also the Area Under the ROC Curve, Matthews Correlation Coefficient, Recall,
Precision, F1 Measure, and others.
 |
Favorite Statistics for all Categories |
GeneXproTools allows you to use Favorite Statistics not only in
the History Panel for model selection but also for ensemble management
in the Deploy Ensemble to Excel Window.
The favorite statistics for Classification & Logistic Regression include:
For Regression & Time Series Prediction the favorite statistics include:
And for Logic Synthesis the favorite statistics include:
 |
Improved History Panel |
The biggest improvement in the History Panel is the implementation of
Favorite Statistics in all categories. But other smaller improvements
were also added, including:
- Random sub-sampling that enables the application of bootstrap techniques to evaluate with
more confidence the accuracy of your models or any
measure of fit;
- Extra functionality accessible both through the context menu and toolbar icons, such as
Delete Active Model and Delete History;
- A more comprehensive summary which now also computes the average of both the favorite statistic and fitness of all the models in the History;
- A new feature for re-indexing all models (Rename All Models) that allows the analysis of models in a particular order;
- The new Add Simple Models functionality that allows the analysis of all input and derived variables as simple models;
- And new functionality for Updating the Rounding Thresholds in Classification and Logistic Regression.
 |
100+ New Fitness Functions |
We’ve added more than one hundred new fitness functions and improved old ones
by combining them with a wider range of adjustable parameters and more sophisticated penalties
for avoiding local optima and strong & mediocre attractors, such as models that classify
everything as zero or one in classification and logistic regression.
Now the vast majority
of GeneXproTools fitness functions combines multiple objectives, such as the use
of different reference simple models in all categories; cost matrix in Classification,
Logistic regression and Logic Synthesis; different types of rounding thresholds
in Classification, including evolvable thresholds; lower and upper bounds for
the model output; parsimony pressure and variable pressure; and many others.
 |
Adjustable Parsimony Pressure for all Fitness Functions |
Parsimony Pressure is now an adjustable parameter that you can fine tune between [0, 1]
in order to apply pressure on the structural complexity of the evolving solutions.
It’s available for all fitness functions, including custom fitness functions.
 |
New and Adjustable Variable Pressure for all Fitness Functions |
Now GeneXproTools also supports Variable Pressure for all fitness functions,
including custom fitness functions. It’s an adjustable parameter between [0, 1],
allowing you to control the blending of variables into your models.
We've also added a new Complexify Button in the Run Panel so that you can easily apply
variable pressure to a particular solution.
 |
More Parameters for the Custom Fitness Functions |
For all modeling categories, we’ve increased the number of pre-computed parameters you can use to design your
own Custom Fitness Functions, including very useful and widely used parameters readily accessible
through the interface, such as the cost matrix, the evolvable rounding thresholds,
model boundaries, and many others.
 |
New Genetic Operators & Modeling Strategies |
We’ve added 20 new genetic operators, covering a wide range of functionalities and
behaviors that you can explore to design your own modeling strategies. From genetic operators
for finding the most effective range for the random constants
(Constant Fine-Tuning & Constant Range Finding),
genetic operators that change only certain structures or elements in the
evolving models (Leaf Mutation, Biased Leaf Mutation, Conservative Mutation,
Conservative Function Mutation, Conservative Permutation, Biased Mutation,
Tail Inversion and Tail Mutation), or genetic operators that inject fresh blood
in the population or increase the frequency of particular models
(Random Chromosomes, Random Cloning and Best Cloning),
you can now have more control over evolution and therefore can push it
in specific directions.
GeneXproTools ships with 4 built-in modeling strategies
that cover some of the most common modeling needs, namely a strategy for
fine-tuning the numerical constants of your models (Constant Fine-Tuning); a strategy for
model fine-tuning where the overall structure of the model remains
basically unchanged making only small changes in the model’s structure (Model Fine-Tuning);
a strategy for Sub-set Selection which is ideal for creating good random forests,
especially for datasets with many variables, as it works only with the elements
that were randomly drawn for the initial population; and of course a
strategy designed for Optimal Evolution which tries to give a good blend
of diversity versus stability in order to optimize and accelerate evolution.
Moreover, GeneXproTools also allows you to create your own modeling strategies by changing any of
the built-in strategies to create a Custom Strategy.
 |
More Variables & Unlimited Ensemble Size |
We’ve extended the number of independent variables in all editions,
now with a max of 20 variables for the Standard Edition, 100 for the
Advanced and 1000 for the Professional.
We’ve also removed all restrictions on ensemble size in all
GeneXproTools editions.
So now you can deploy ensembles to Excel of any size you need.
 |
New Tools for Creating Ensembles & Random Forests |
We've introduced new tools for creating ensemble
models or GEP forests, including new fitness functions,
new genetic operators, subset selection strategies, favorite statistics,
unlimited ensemble size for external
ensemble deployment to Excel, ensemble deployment in data-only mode,
average probability multi-model & median probability multi-model
for Classification and Logistic Regression,
different bagging schemes and different stop conditions.
 |
New Programming Languages: R, Octave & Excel VBA |
We've now added 3 new grammars to the already extensive list of supported programming languages
for automatic model code generation.
The R Language, Octave and Excel VBA are now part of the
19 built-in grammars of GeneXproTools
(Ada, C, C++, C#, Excel VBA, Fortran, Java, JavaScript, Matlab, Octave, Pascal, Perl,
PHP, Python, R, Visual Basic, VB.Net, Verilog, and VHDL)
that allow you to export automatically all your models to any of these programming languages.
 |
Improvements in Generated Model Code |
We’ve changed the implementation of functions involving the ternary operator
in order to avoid very long lines of code, implementing these functions as
individual methods. We’ve also improved the grammars, declaring only the
numerical constants and variables that are being used in the model,
improving the clarity of the model code.
 |
3 Different Forms of Model Output for Classification & Logistic Regression |
We’ve now implemented throughout all GeneXproTools panels support for 3 different forms
of mode output for classification and logistic regression models, namely
Probability[1],
Most Likely Class and Raw Model Output.
Internally GeneXproTools gives you access to these 3 different forms of model output in
the Data Panel, for both classification and logistic regression models.
In the Results Panel and for Logistic Regression, you also have access to
the 3 different forms
of model output. For Classification, however, GeneXproTools
shows only the raw model output and the predicted class in the Results Panel. The
probabilities for classification models are available in the
Data Panel and through model scoring,
either in the Scoring Panel or through model/ensemble deployment to Excel.
For scoring your models internally in GeneXproTools
in the Scoring Panel, you can also choose
any of these 3 forms
of model output for classification and logistic regression models. This is also true
for Model & Ensemble Deployment to Excel.
Finally, all the code generated by GeneXproTools in any of the supported
programming languages for mathematical models (Ada, C, C++, C#, Excel VBA, Fortran, Java, JavaScript, Matlab, Octave, Pascal, Perl,
PHP, Python, R, Visual Basic, and VB.Net), implements also the 3 different forms of model output:
Probability[1], Most Likely Class and Raw Model Output, which you select directly
in the Model Panel.
 |
Ensemble Deployment to Excel with Average & Median Probability Models |
Now for Logistic Regression and Classification, you can choose Probability[1]
as the output for your models, both for ensemble and model deployment to Excel.
For ensemble deployment we’ve added the new average probability model and
median probability model, with the thresholds for the probabilities easily
adjustable within Excel.
 |
Ensemble Deployment without Embedded Code |
Now GeneXproTools also supports ensemble and model deployment to Excel in
data-only mode, that is, without
embedding the model code. This is especially useful if you are dealing with
large ensembles and large datasets, particularly in the ensemble evaluation phase
when you need to quickly evaluate different candidate solutions.
 |
New Linking Functions |
We’ve now added 3 more linking functions: Avg2, Min and Max, all of which
can be easily added to your own grammars. The Avg2 linking function works
particularly well with time series data, producing better models by
preventing autocorrelation, and therefore is now the new
default in Time Series Prediction.
 |
Improved Expression Tree Display |
Now you can access the labels of all the variables in the expression trees with
the help of tooltips. GeneXproTools also displays the definitions of all the functions and
the values of the numerical constants in the tree representation of the model code.
 |
New Defaults for the Function Sets |
We created new defaults for the functions sets of all categories in an attempt
to create an even better starting point for as wide a range of problem domains as possible.
We’ve also added new functionality to the Functions Panel so that
you can now access all these different
function set defaults from different modeling categories.
 |
Import Function Set |
We’ve introduced a new feature for importing function sets from other gep files.
This also includes importing the code of all custom functions designed for a particular run.
 |
Import Derived Variables |
GeneXproTools allows you to import the code of all the
new derived variables you’ve created.
So now you no longer have to copy and paste the code of your favorite derived variables or
variable transformations from one run to another. You just have to write them once and
easily access them through the Import Derived Variables menu.
 |
Analysis of Simple Models |
GeneXproTools now allows you to Add Simple Models to the History so that
you can analyze and also compare them to your models. The simple models
you can add automatically to the History include all the original variables
in your data and all the derived variables you've created.
 |
New Charts for Monitoring Evolution |
In the Run Panel GeneXproTools now shows the histograms of the fitness values and
program sizes for each generation of evolving models. This is particularly useful
if you are designing your own fitness functions or creating your own modeling strategies
by adjusting the rates of the genetic operators.
 |
New Stop Conditions |
We’ve introduced 6 new Stop Conditions, some of which are very useful
for creating good ensembles or random forests, such as
Random Generation Number,
Random Fitness Threshold, Random R-square Threshold and
Random Accuracy Threshold.
These stop conditions allow you to choose the limits for the lower and upper value for the
number of generations, fitness, R-square or accuracy, allowing you to stop the design process
at different stages, thus allowing you to create much more diverse ensembles of models.
 |
New Online Help System |
We’ve now introduced a new Online Help System, providing you with constantly
updated materials that we will be updating and improving in response to users needs.
 |
And Much More… |
The list of smaller improvements and features added to GeneXproTools 5.0 is very large,
but below you’ll find the most salient of them.
Improved Classification Tapestry
Now, if no models exist in the History, the new Classification Tapestry shows
simply the distribution of Positives and Negatives in the response variable.
The new Classification Tapestry also uses another color scheme when the accuracy
is below the 50% mark, which can happen not only for weak intermediate solutions
but also for good inverted models
when
fitness functions
that select for both positive
and negative correlations are used. The
R-square and
Symmetric ROC fitness functions
of Logistic Regression are examples of such
fitness functions.
Improved All Curve Fitting Charts
All the Curve Fitting Charts of Regression and Time Series Prediction were improved
with a better design and by adding extra functionality, such as zooming, showing labels,
legend, titles, tooltips and gridlines.
Improved All Binomial Fitting Charts
The old Binomial Fitting Charts of Logistic Regression
were improved with a better design and by adding extra functionality, of which
the most important is plotting the rounding threshold and
highlighting the misclassifications. The new ones (all three Binomial Fitting Charts of
Classification and the brand new Binomial Fitting Chart Sorted by Target & Model) were
also created with similar design and features.
Improved Logistic Regression Window Charts
The charts for Quantile Regression, Cutoff Points, Log Odds and Logistic Fit
of the Logistic Regression Window were also improved and redesigned.
Conversion of Logistic Regression Runs to Classification
Now, thanks to the introduction of
evolvable rounding thresholds,
the conversion of Logistic Regression runs to Classification is done
flawlessly for all the models in the Run History. By setting the
rounding threshold to Logistic Thresholds, it is now possible to bring the
unique logistic threshold of each logistic regression model to the Classification Platform.
Improved Change Seed Window
Now in the Change Seed Window you can copy & paste the entire set of
numerical constants at once or copy & paste just the numerical constants
of a particular gene.
New Complexify Button in the Run Panel
Variable Pressure was introduced in version 5.0 and we also added a
Complexify Button to the Run Panel, giving you an easy way of controlling
the blending of variables into your models.
New Modalities for the Report Panel
Now GeneXproTools allows you to choose between a Full Report and
a Short Report. The Full Report shows a list of all the models in the Run History,
whereas the Short Report lists only the active model. Choosing the shorter version is
advisable for very large histories with hundreds or thousands of models.
Improved Notes
We’ve improved the Notes Editor of GeneXproTools with a larger and
better window and also provided easier access to the Notes Editor through the Reports Menu.
New Code Editor for the Custom Fitness
We’ve improved the Code Editor for the Custom Fitness Function through a
larger and better window and the use of different colors for JavaScript keywords,
comments and constants.
Improved User Defined Functions Window
We’ve improved the Dynamic UDFs Window with a new combobox for the
number of arguments and a new Code Editor that now allows the use of
different colors for JavaScript keywords, comments and constants.
Improved Derived Variables Window
We’ve improved the UDFs Window, which is the window for generating new
derived variables, with a new Code Editor that now highlights in
different colors all JavaScript keywords, comments and constants. We’ve also added
new functionality for copying the output of each test.
Extra Functionality in the Functions Panel
Now the Select All and Clear buttons work more intelligently,
selecting/clearing only the functions that are visible.
We’ve also added a new feature that allows you to copy all the sub-sets of functions
shown in the Functions Table, which is useful for designing and studying your own function sets.
New Home Panel
We’ve redesigned the Home Panel of GeneXproTools, giving you access
to a larger list of the most recent runs. We’ve also added a list of
useful links for getting help and support and easy access to the new
discussion forum.
New System for Identifying Modeling Categories
Now in GeneXproTools 5.0 we color-code each modeling category to help in the
quick identification of the modeling category of each run and to make everything more,
well, colorful.
New Sample Runs
We’ve created new Sample Runs for this version so that you could
easily explore all the new features of GeneXproTools 5.0, such as the
support for categorical variables and missing values, the support for
multiple classes in Classification and Logistic Regression, data normalization,
sub-sampling, and so on.
For example, the Classification sample run Iris Plants has 3 classes (Iris Setosa,
Iris Versicolor and Iris Virginica),
allowing you to explore with ease how to handle multiple classes in Classification.
The Satellite Images sample run plays a similar role for Logistic Regression,
with 6 classes in this case. The Credit Approval sample run deals with
standardized data. Some datasets of the new sample runs have missing values
(Credit Approval, Breast Cancer, Diabetes and Fuel Consumption), while others use a
mix of categorical and numerical variables (Credit Approval, Breast Cancer,
Iris Plants, Loan Risk, Satellite Images and Emotiv EEG).
For all benchmark sample runs
that deal with well-known datasets, we used the original datasets as provided
by the donors so that you could compare and reproduce any other studies
conducted using these datasets. To revert to the original dataset,
just press the Original Button in the Dataset Partitioning Window.
New Defaults for the Genetic Operators
With so many new
genetic operators we had to readjust their rates to make the best of evolution.
Moreover, now all genetic operators are implemented probabilistically, so now you can use
very small rates and still get them to work probabilistically
over extended periods of time.
And finally, each of the new
modeling strategies comes with specific defaults that
you can access every time you choose a particular strategy.
Improved Evolutionary Dynamics Chart
The chart for monitoring the evolutionary dynamics in the Run Panel was
improved with a better design and by adding support for zooming, labels, legend,
titles and better tooltips.
Improved Average/Best Size Chart
The chart for visualizing the changes in Best Size and Average Size during evolution
was improved with a better design and by adding support for zooming, labels, legend,
titles and better tooltips.
Improved Sub-Program Sizes Chart
We’ve improved the Sub-Program Sizes Chart in the Run Panel with a better design and
by adding extra functionality for labels and copying the chart data.
Improved All Sizes Chart & All Fitnesses Chart
We’ve also improved the All Sizes Chart and All Fitnesses Chart in the Run Panel
with a better design and by adding support for labels and copying the chart data.
Improved Navigation of All Charts
We’ve now improved the navigation of all charts in the Run Panel and
Results Panel through the use of arrows that work in concert with a
particular combobox. So now it’s much easier to go back and forth between
different charts both during evolution in the Run Panel and during model analysis
in the Results Panel.
New Sorting Tools in the Results Panel
We’ve now added Record IDs to all Results Tables and extended the
sorting options in all modeling categories.
More Copy Options for the Results Tables
We’ve extended the number of copying options for the Results Table in
all modeling categories in order to include all the new additions to the Results Tables,
such as Copy Probability[1] in Logistic Regression.
New Custom Fitness Functions Examples
Now the Custom Fitness Functions examples for
Classification,
Logistic Regression
and Logic Synthesis are implementations of a simple fitness function based on the
Classification Accuracy. For
Regression and
Time Series Prediction we now implement
a fitness function based on the RMSE.
New DDFs Examples
We created new examples for the math and Boolean custom functions in order to provide
more useful examples that are more interesting to use.
Improved Defaults
We've improved a few defaults for things to run even more smoothly:
- We've changed the Function Set defaults in all modeling categories
in order to create even more accurate models and to work well with the
new genetic operators;
- In Time Series Prediction we've changed significantly
the Function Set default in order to make the best of the
new Avg2 linking function default;
- Now all fitness functions with adjustable parameters come with
preset defaults to help you make the most of them without the need for
understanding their inner workings in all detail. These defaults reset
every time you change the fitness function so that you always have a
good starting point.
Release Date: May 28, 2013
Legacy:
4.3 |
4.0
|





