| The successful creation of Classification models requires the use of very sharp modeling
tools and the most important are the following:
Data Screening Engine
GeneXproTools
accepts two different data formats: the standard Samples x
Variables format where samples are in rows and variables in
columns, with the class occupying the
rightmost position; and Gene Expression Matrixes commonly used
in DNA microarray studies where samples are in columns and
variables in rows, with the class occupying the topmost position. Note,
however, that GeneXproTools uses
the Samples x
Variables format and therefore all formats are automatically
converted and shown in this format.
The data screening engine of GeneXproTools 4.0 checks the validity of all the data sets used both in the creation and testing of the model.
The data screening engine is operative every time you load data sets
either for training or testing.
Classification problems require data files with numerical inputs in which the dependent variable can take only two values: zero or one. The data screening engine of
GeneXproTools makes sure that all the input files for classification respect this format. But it is common for the input files to have missing or nominal values which render them useless for modeling or testing.
GeneXproTools 4.0 data screening engine can help you identify these faulty samples and correct them.
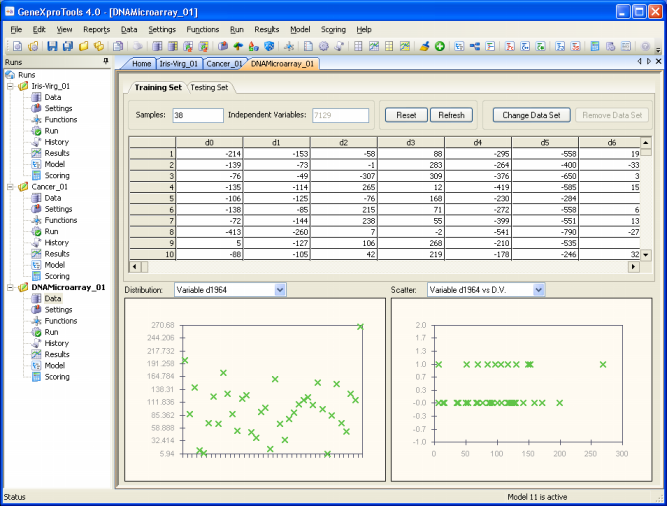
Data Visualization Tool
The data visualization tool of GeneXproTools 4.0 enables you to analyze the distribution of all the inputs and to spot simple relationships between the independent variables and the dependent variable.
The visualization of both the distribution and relationships between variables is a valuable modeling tool as it can help you detect outliers or simple relationships between your variables that can be used to evolve more efficient models.
Built-in Fitness Functions
GeneXproTools 4.0 offers 30 built-in fitness functions based on well-known statistical functions.
Fitness functions are fundamental modeling tools for they determine the nature of the search landscape. And different fitness functions open and explore different paths in the solution space. The
30 built-in fitness functions of
GeneXproTools 4.0 are named as follows: Number of Hits,
Hits with
Penalty, Accuracy,
Squared Accuracy,
Sensitivity/Specificity,
PPV/NPV,
SSPN, R-square,
Correlation Coefficient, MSE, RMSE,
MAE, RSE,
RRSE, and RAE.
Furthermore, all these fitness functions have their own counterpart
with parsimony pressure.
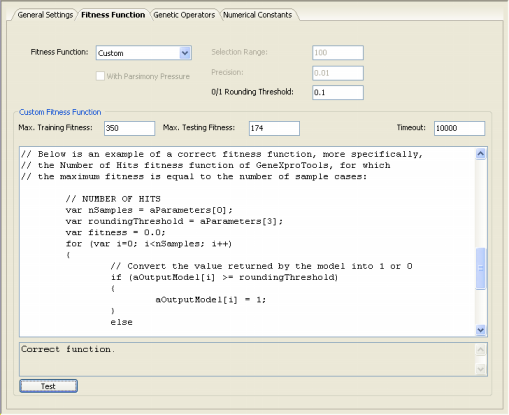
User Defined Fitness Functions
Despite the wide set of GeneXproTools built-in fitness functions, some users sometimes want to experiment with fitness functions of their own.
GeneXproTools 4.0 gives the user the possibility of creating custom tailored fitness functions and evolve models with them.
The code for the custom fitness function must be in JavaScript and is written in the Custom Fitness Function window and can be tested before evolving a model with it.
Built-in Mathematical Functions
GeneXproTools 4.0 offers a total of 279 built-in mathematical
functions, including 186 different IF THEN ELSE rules. This wide set of functions allows the evolution of complex and rigorous models quickly built with the most appropriate mathematical functions or rules.
From the simple arithmetic operators to complex mathematical functions available in most programming languages to more complex mathematical functions commonly used by engineers and scientists, the modeling algorithms of
GeneXproTools 4.0 enable you the integration of the most appropriate functions or rules in your models.
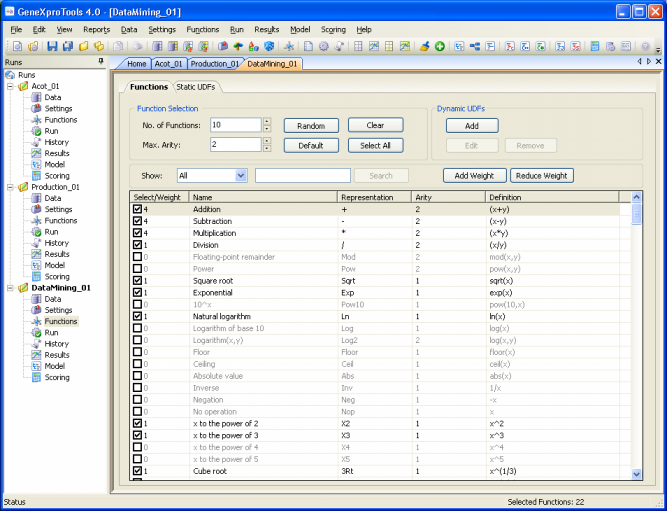
Function Selection Tool
For some problems, the simple arithmetic operators are more than
enough to create extremely accurate and elegant models. But for
other problems more complex functions are necessary to create good
models and when inside knowledge of the problem at hand is not
producing the expected results, it might be interesting to
experiment with different combinations of functions. The Function
Selection Tool of GeneXproTools 4.0 helps you to experiment with
different function sets very quickly either by pressing the Random
button in the Functions Panel or by selecting certain sub-sets of
functions.
In addition, although you can always handcraft your function set so
that it is well-balanced relatively to the number of independent
variables in your data, in GeneXproTools 4.0 this is no longer
essential as GeneXproTools balances automatically all function sets.
This is particularly important for high-dimensional datasets, such
as the huge DNA microarray data.
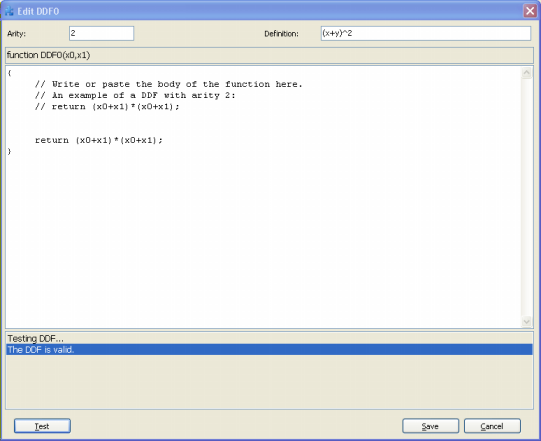
Dynamic User Defined Functions (DDFs)
Despite the wide set of GeneXproTools built-in mathematical functions,
some users sometimes want to model with different functions or rules.
GeneXproTools 4.0 gives the user the possibility of creating custom tailored functions and evolve models with them. A note of
caution though: the use of DDFs slows considerably the evolutionary process and therefore should be used with moderation.
The code for the DDFs must be in JavaScript and is written in the Edit DDF window (in the Functions Panel, select the Functions Tab and then click Add in the Dynamic UDFs tool box).
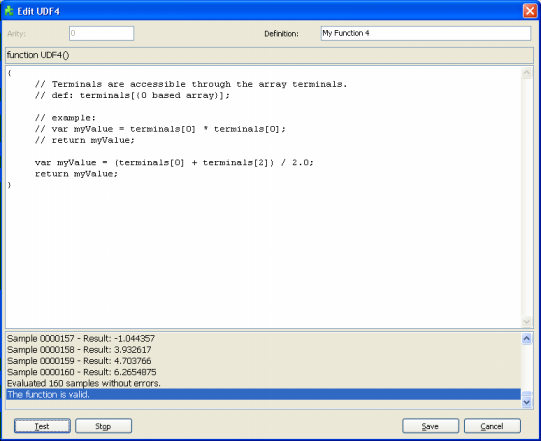
Static User Defined Functions (UDFs)
Sometimes it is possible to spot relatively simple relationships between certain variables in your data, and
GeneXproTools 4.0 allows you to use this information to build custom tailored functions (UDFs). These functions allow the discovery of more complex models composed of several simpler models.
The code for the UDFs must be in JavaScript and is written in the Edit UDF window (in the Functions Panel, select the Static UDFs Tab and then click Add in the Static UDFs tool box).
Learning Algorithms
GeneXproTools 4.0 is equipped with two different learning
algorithms for evolving complex nonlinear classification models.
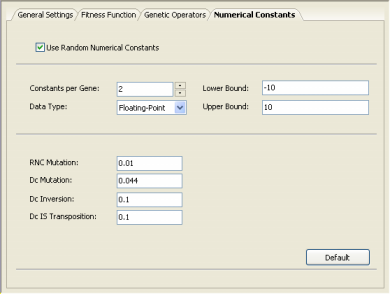
The first is the fastest and simplest of the two. The second algorithm is very similar to the first, with the difference that it gives the user the possibility of choosing the range and type of numerical constants that will be used during the learning process.
And because numerical constants are crucial to a wide set of
problems, the second algorithm is the default in
GeneXproTools 4.0. Keep in mind, however, that for problems where
numerical constants are not necessary it pays to use the first
algorithm as the models are not only much simpler but also the
evolutionary process is much quicker.
In order to evolve models using the second algorithm, select Numerical Constants in the Settings Panel and then check the box that activates this algorithm. You will notice that additional parameters become available, including a small set of genetic operators especially developed for handling numerical constants (if you are not familiar with these operators, please use the default values for they work very well in all cases).
To use the first algorithm, just uncheck the Use Random Numerical
Constants box.
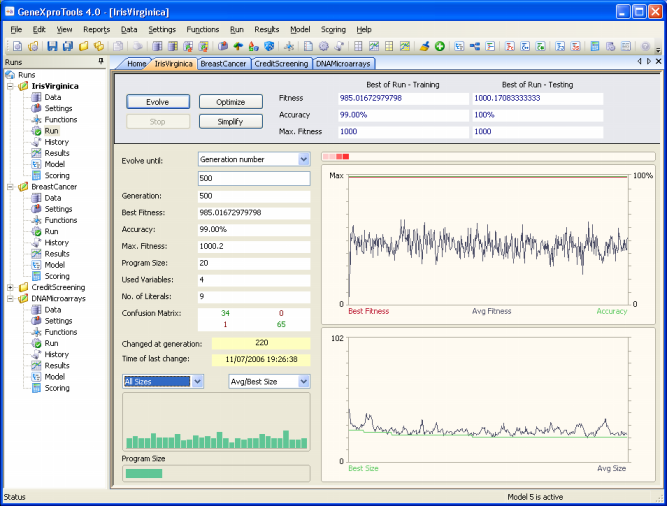
Monitoring the
Modeling Process
GeneXproTools 4.0 allows you to monitor the modeling
process by showing the essential parameters of a run during the discovery process, including the fitness,
classification accuracy,
and confusion matrix (true positives, true negatives, false positives, and false negatives) of the best-of-generation model.
Furthermore, GeneXproTools allows you to visualize the modeling
process by plotting the essential features of a run during the discovery
process. In the Run
Panel, a total of eight different charts are
shown:
- The Variables Usage map.
This map shows not only the variables that are being used by the
best-of-generation models but also their weight. Not only after
a run but also during evolution, by placing the cursor over each
square you can access the variable ID and the number of times it
appears in the current model. And by clicking the right button
of your mouse, you can also change its appearance: Heat Map,
Random Colors, or Monochromatic.
- The Evolutionary Dynamics chart.
This chart shows
the average fitness of the population plus the fitness and accuracy of the best-of-generation model.
- The Average/Best Size chart.
This chart shows
the average program size of the population versus the program size of the best-of-generation model.
This chart is especially useful during simplification
and can be activated any time during evolution by selecting Avg/Best
Size in the rightmost combo box in the bottom.
- The Target/Model chart.
This chart plots the first 50 data points of the training set
and shows how well the evolving models are fitting the target.
The Target/Model chart can be activated any time during
evolution by selecting Target/Model in the rightmost combo box
in the bottom.
- The Sub-Program Sizes chart.
This chart shows
the sizes of all the sub-programs in the best-of-generation model.
Not only after a run but also during evolution, by placing the
cursor over each bar you can access the size of all sub-programs
in your model. The Sub-Program Sizes chart can be activated any
time during evolution by selecting Sub-Program Sizes in the
leftmost combo box in the bottom.
- The All Sizes chart.
This chart shows
the sizes of all the models in the population. Not only after a run
but also during evolution, by placing the cursor over each bar
you can access the size of a particular model; the
best-of-generation model always occupies the first position so
you can also easily see how it fares relatively to the others.
The All Sizes chart can be activated any time during evolution
by selecting All Sizes in the leftmost combo box in the bottom.
- The All Fitnesses chart.
This chart shows
the fitnesses of all the models in the population. Not only after a
run but also during evolution, by placing the cursor over each
bar you can access the fitness of a particular model; the
best-of-generation model always occupies the first position so
you can also easily see how it fares relatively to the others.
The All Fitnesses chart can be activated any time during
evolution by selecting All Fitnesses in the leftmost combo box
in the bottom.
- The Program Size chart.
This chart shows
the size of the best-of-generation model. Not only after a run but
also during evolution, by placing the cursor over the horizontal
bar you can access the size of the best model.
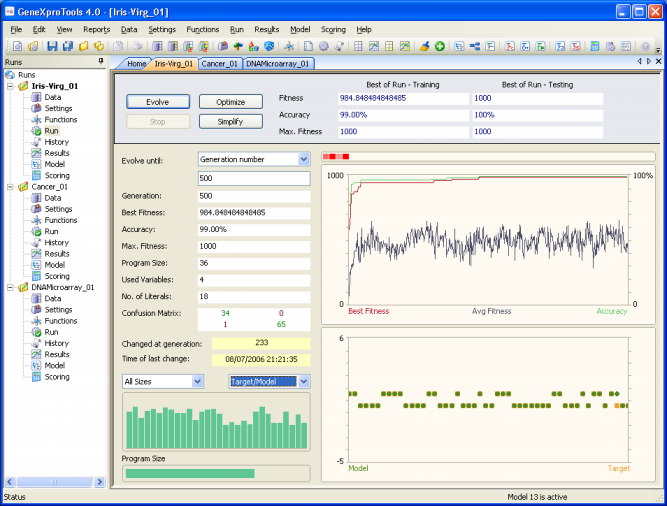
The evolutionary process can be stopped whenever you are satisfied with the results by pressing the Stop button or you can use one of the stop criteria of
GeneXproTools for picking up your model exactly as you want it.
When the evolutionary process stops, the best-of-run model is ready either for analysis or scoring. And if you are still not happy with the results, you can still try to fine-tune the evolved model by pressing the Optimize button and repeat this process until you are completely satisfied with your model.
In addition, you might also wish to try and simplify your model and
for that you just have to press the Simplify button.
Analyzing Intermediate Models
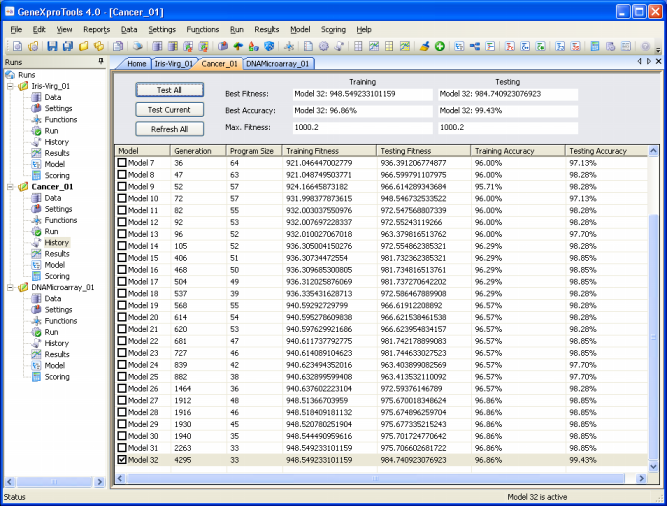
GeneXproTools 4.0 gives you the possibility of gaining some insights into the modeling process by analyzing all the best-of-generation intermediate models.
All the best-of-generation models discovered during a run are saved and you can pick them up for analysis at the
History Panel by checking the model you are interested in. Each of these intermediate models can then be analyzed exactly as you do for the best-of-run model, that is, you can check their performance in the testing set, check their vital statistics, automatically generate code with them, visualize their parse trees, use them as seed to create better models from them and so forth.
Simplifying a Model
GeneXproTools 4.0 allows you to simplify an existing model (which could have been either generated by
GeneXproTools or by another modeling tool) either by pressing the
Simplify button on the Run Panel or by choosing a fitness function
with parsimony pressure and then press Optimize or Simplify on the
Run Panel.
For models created outside of GeneXproTools or for GeneXproTools
models modified by the user, the starting model is fed to the algorithm through the Change Seed window where both the fitness and structural soundness of the model are tested.
Then, in the Run Panel, by clicking the
Simplify button, an evolutionary process starts in which all the subsequent models will be descendants of the
model you want to simplify. Keep in mind, however, that the
simplification algorithm used by GeneXproTools is an evolutionary
one and, despite parsimony pressure being applied, models continue
to be selected preferentially by fitness and sometimes their
complexity might even increase temporarily if the gain in fitness outweighs
the loss in simplicity.
For models created in the GeneXproTools 4.0 environment, you just
have to select the model you want to simplify (either the
best-of-run or an intermediate model picked up in the History Panel)
and then click the Simplify button and let the algorithm create
better descendants not only in terms of fitness but also in terms of
size.
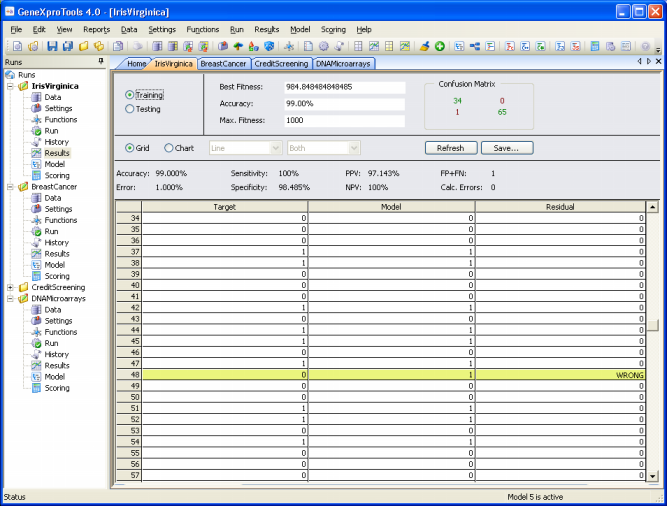
Comparing Actual and Predicted Values
GeneXproTools 4.0 offers two different ways of analyzing and comparing the output of your model with the actual or target values both for the training and testing sets.
In the first, the target or actual values are listed in a table side by side with the predicted
or model values.
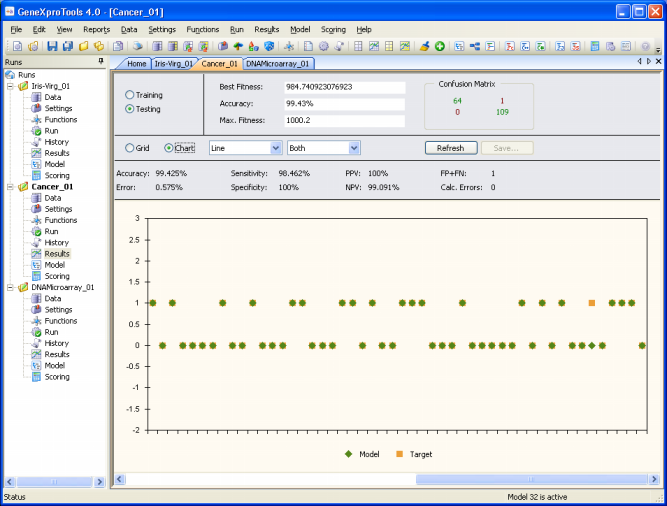
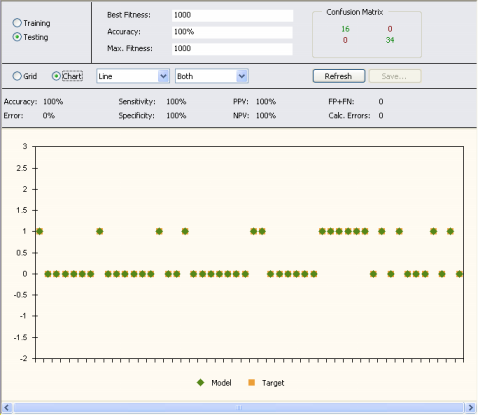
In the second, the target and predicted values are plotted in a
Target/Model chart for easy visualization. By moving the scroll bar
in the bottom, you can see the results for the entire dataset.
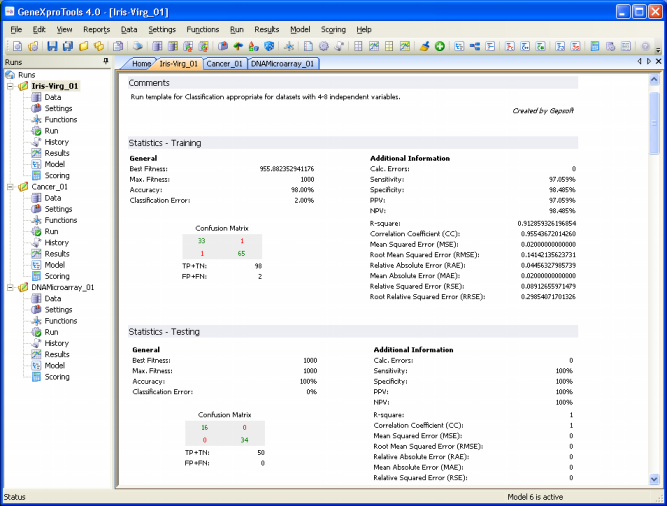
Essential Statistics
GeneXproTools 4.0 allows a quick and easy assessment of a wide set of
statistical
functions. Some of them are immediately computed and shown every time you check the
Results Panel
(classification error,
classification accuracy,
confusion matrix (true positives, true negatives, false positives, and false negatives),
sensitivity, specificity,
positive predictive
value, and negative predictive
value).
Others, tough, are only shown in the Report Panel (mean squared
error, root mean squared
error, mean absolute
error, relative squared
error, root relative squared
error, relative absolute
error, R-square,
and correlation
coefficient) after their evaluation in the Results Panel.
Modeling from Seed
GeneXproTools 4.0 allows the use of an existing model (which could have been either generated by
GeneXproTools or by another modeling tool) as the starting point of an evolutionary process so that more complex, finely tuned models could be created.
For models created outside of GeneXproTools or for GeneXproTools
models modified by the user, the starting model or seed is fed to the algorithm through the Change Seed window where both the fitness and structural soundness of the model are tested.
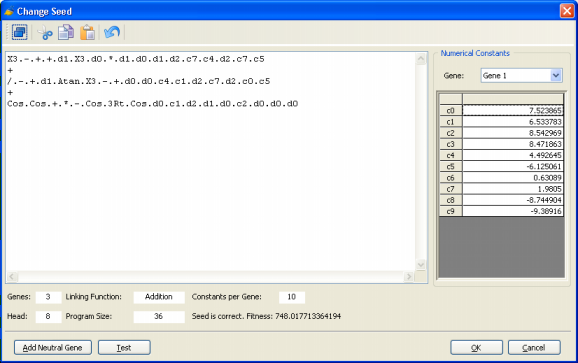
Then, in the Run Panel, by clicking the Optimize button, an evolutionary process starts in which all the subsequent models will be descendants of the seed you introduced. A note of caution though: if your seed has a very small fitness, you risk losing it early in the run as better models could be randomly created by
GeneXproTools and your seed would, most probably, not be selected for breeding new models. If your seed has zero fitness, though, you will receive a warning so that you could
modify your seed until it becomes a viable seed capable of breeding new models.
For models created in the GeneXproTools 4.0 environment, the seed is fed to the algorithm every time you click the Optimize
or Simplify buttons during modeling, that is, the seed (in this case, the best model of the previous run)
is introduced automatically by
GeneXproTools.

Adding a Neutral Gene
The addition of a neutral gene to a seed might seem, at first sight, the wrong thing to do as most of the times we are interested in creating efficient and parsimonious models. But one should look at this as modeling in progress as, for really complex phenomena, it is not uncommon to approximate a complex function progressively.
Thus, being able of introducing extra terms in your seed is a powerful modeling tool and
GeneXproTools 4.0 allows you to do that by selecting Add Neutral
Gene in the Edit Menu or through the Change Seed window.

Here, by pressing the Add Neutral Gene button, you will see a neutral gene being added to your model. By doing this, you are giving the learning algorithm more room to play and, hopefully, a
better and more complex program will evolve.
Neutral genes can also be introduced automatically by GeneXproTools 4.0
using the Complexity Increase
Engine.
Complexity Increase Engine
GeneXproTools 4.0 also allows you to introduce neutral genes automatically by activating the Complexity Increase
Engine in the Settings Panel -> General Settings Tab.
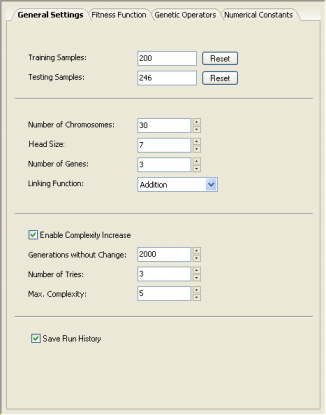
Whenever you are using the Complexity Increase engine of GeneXproTools 4.0, you must fill the Generations Without Change box to set the period of time you think acceptable for evolution to occur without improvement in best fitness, after which
a mass extinction or a neutral gene (an extra term) is automatically added to your model; the Number of Tries box corresponds to the
number of consecutive evolutionary epochs (defined by the parameter
Generations Without Change) you will allow before a neutral gene is
introduced in all evolving models; in the Max. Complexity box you write the maximum number of terms (genes) you’ll allow in your model and no other terms will be introduced beyond this threshold.
The Complexity Increase Engine of GeneXproTools 4.0 might be a very powerful modeling tool, but you must be careful not to create excessively complex models for, most of the times, a greater complexity does not
necessarily imply a greater efficiency.
Visualizing Models as Parse Trees
GeneXproTools 4.0 comes equipped with a parse tree generator that automatically converts your models into diagram representations or parse trees.

Indeed, all the models evolved by GeneXproTools in its native Karva language can be automatically parsed into visually appealing expression trees, allowing a quicker and more complete understanding of their mathematical intricacies.
Generating Code Automatically Using GeneXproTools Built-in Grammars
GeneXproTools 4.0 offers a total of 15 built-in grammars so that the models evolved by
GeneXproTools in its native Karva language
can be automatically translated into the most commonly used programming languages
(Ada, C, C++, C#, Fortran, Java, Java Script, Matlab, Pascal, Perl, PHP, Python, Visual Basic, VB.Net, and VHDL). This code can then be used in other applications.
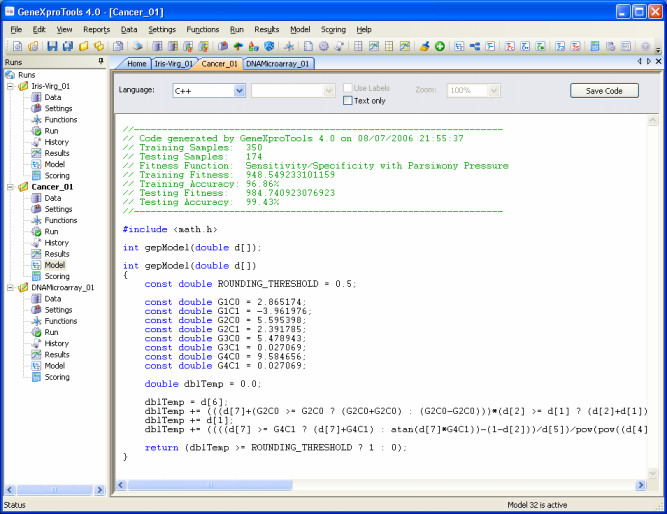
Generating Code Automatically Using User Defined Grammars
GeneXproTools 4.0 allows the design of User Defined Grammars so that the models evolved by
GeneXproTools in its native Karva language
can be automatically translated into the programming language of your choice if you happen to prefer a programming language not already covered by the
15 built-in grammars of
GeneXproTools 4.0 (Ada, C, C++, C#, Fortran, Java, Java Script, Matlab, Pascal, Perl, PHP, Python, Visual Basic, VB.Net, and VHDL).
As an illustration, the C++ grammar of GeneXproTools 4.0 is shown here.
Other grammars may be easily created using this or other GeneXproTools
built-in grammars as reference.
Scoring Engine
GeneXproTools 4.0 comes equipped with a scoring engine for scoring your databases/Excel or text files in the
GeneXproTools environment. The scoring engine of GeneXproTools uses
the JavaScript code of your models to score your data.
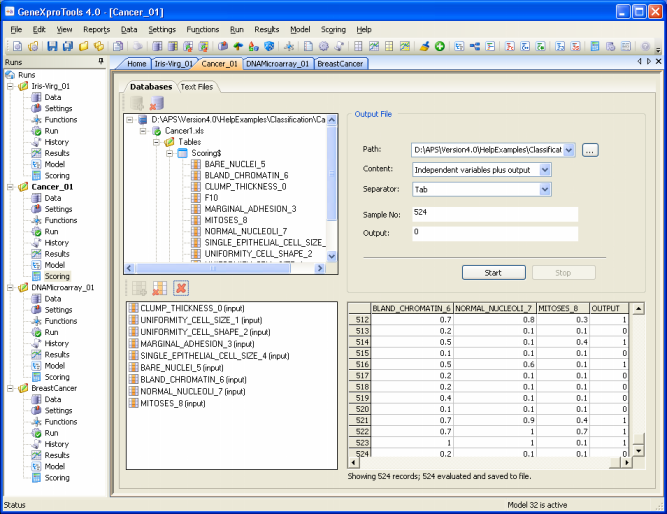
After its creation, a model is usually used multiple times to make predictions or extrapolations. The scoring engine of
GeneXproTools 4.0 allows the immediate scoring of your databases as soon as a model is created, saving the scores to
Excel (csv) or text files.
|

















