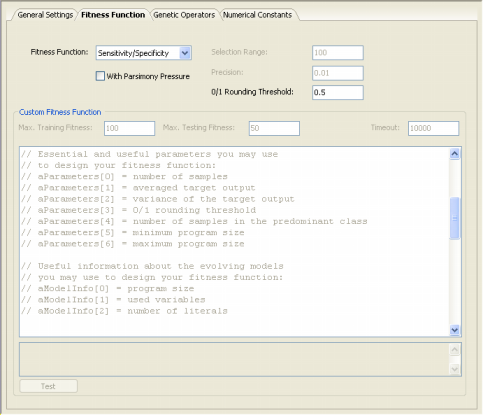
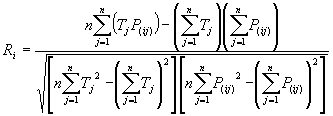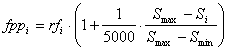| GeneXproTools 4.0 implements the R-square fitness function both with and
without parsimony pressure. The
version with parsimony
pressure puts a little pressure on the size of the evolving
solutions, allowing the discovery of more compact models. For all classification problems, in order to be able to apply a particular fitness function, the learning algorithms of GeneXproTools 4.0 must convert the value returned by the evolved model into “1” or “0” using the 0/1 Rounding Threshold. If the value returned by the evolved model is equal to or greater than the rounding threshold, then the record is classified as “1”, “0” otherwise. Thus, the 0/1 Rounding Threshold is an integral part of all fitness functions used for classification and must be appropriately set in the Settings Panel -> Fitness Function Tab.
The R-square fitness function is, as expected, based on the standard
R-square, which returns the square of the Pearson product moment correlation coefficient.
where P(ij) is the value predicted by the individual program
i for fitness case j (out of n fitness cases);
and Tj is the target value for fitness case j. fi = 1000*Ri*Ri and therefore ranges from 0 to 1000, with 1000 corresponding to the ideal.
where Si is the size of the program, Smax and Smin represent, respectively, maximum and minimum program sizes and are evaluated by the formulas: Smax = G (h + t) Smin = G where G is the number of genes, and h and t are the head and tail sizes (note that, for simplicity, the linking function was not taken into account). Thus, when rfi = rfmax and Si = Smin (highly improbable, though, as this can only happen for very simple functions as this means that all the sub-ETs are composed of just one node), fppi = fppmax, with fppmax evaluated by the formula:
|