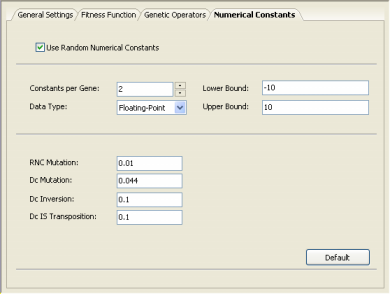
| GeneXproTools 4.0 uses two different learning algorithms for
Time Series Prediction problems. The first – the basic gene expression algorithm
or simply Gene
Expression Programming (GEP) – does not support the direct manipulation of random numerical constants,
whereas the second – GEP
with Random Numerical Constants or GEP-RNC
for short – has a facility for handling them directly. So, these
two algorithms search the solution landscape differently and
therefore you might wish to try them both on your problems. The kinds of models these algorithms produce are quite different and, when both of them perform equally well on the problem at hand, you might still prefer one to the other. But there are cases, however, where numerical constants are crucial for an efficient modeling and, therefore, the second algorithm is the default in GeneXproTools 4.0. You activate this algorithm in the Settings Panel -> Numerical Constants by checking the Use Random Numerical Constants box.
The GEP-RNC algorithm is slightly more complex than GEP
as it uses an additional gene domain (Dc) for encoding the random
numerical constants. Consequently, this algorithm comes equipped
with an additional set of genetic operators (RNC
mutation, Dc mutation,
Dc inversion, and Dc
IS transposition) especially developed for handling random
numerical constants (if you are not familiar with these operators,
please use the default values by clicking the Default button for
they work very well in all cases). |