| The chromosome architecture of your models include the
head size, the number of genes and the linking
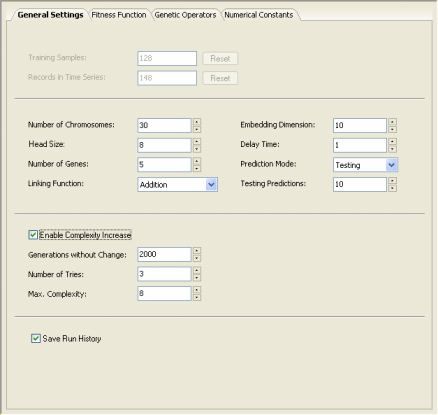
function. You choose these parameters in the Settings Panel -> General Settings.
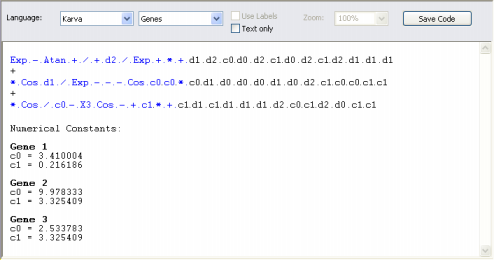
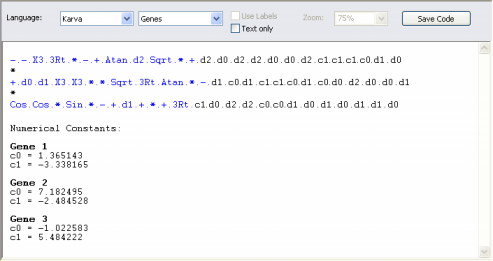
The Head Size determines the complexity of each term in your model. In the heads of genes, the GeneXproTools learning algorithm tries out different arrangements of functions and terminals (variables and constants) in order to model your data. The plasticity of this architecture allows the discovery of a virtually infinite number of models of different sizes and shapes which are afterwards tested and selected during the learning process. The heads of genes are shown in blue in the compact, linear representation of your models in the Model Panel.
More specifically, the head size h of each gene determines the maximum width w and maximum depth d of the sub-expression trees encoded in the gene, which are given by the formulas: w = (n - 1) * h + 1 d = ((h + 1) / m) * ((m + 1) / 2)
where m is minimum arity and n is maximum arity.
The number of genes per chromosome is also an important parameter. It will determine the number of (complex) terms in your model as each gene codes for a different
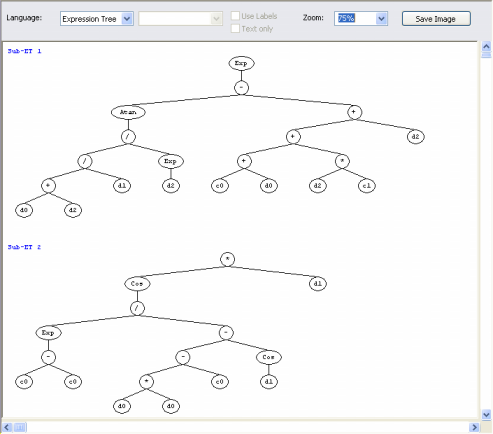
parse tree (sub-expression tree or sub-ET). Theoretically, one could just use a huge single gene in order to evolve very complex models. But the partition of the chromosome into simpler, more manageable units gives an edge to the learning process and more efficient and elegant models can be discovered using
multigenic chromosomes.
|