Automatic Problem Solver 3.0 uses two different learning
algorithms for Time Series Prediction problems. The first is the
most efficient in evolutionary terms as it uses simpler chromosomal
architectures and therefore can discover very good models in record
time. Fortunately enough, this algorithm is also the most efficient
in terms of the quality of the models created. For obvious reasons,
this algorithm – the basic
gene expression algorithm – is the default in APS 3.0.
The chromosomal architecture
of the basic gene expression algorithm does not support the direct
manipulation of random numerical constants and, therefore, it can
only create numerical constants from scratch or invent new ways of
representing them.
The second learning
algorithm of APS 3.0 explores a different chromosomal
architecture that allows the direct manipulation of random
numerical constants and, therefore, can be used to design complex
models with more conventional tools. You activate this algorithm in
the Settings Panel -> Numerical Constants by checking the Use
Random Numerical Constants box.
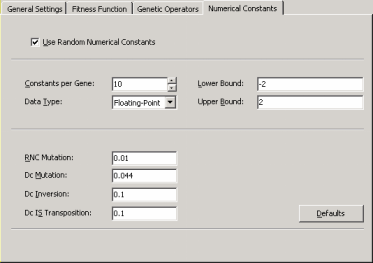
As mentioned above, the second learning algorithm – gene
expression programming with random numerical constants or GEP-RNC
for short – is slightly more complex than GEP
as it uses an additional gene domain (Dc) for encoding the random
numerical constants. Consequently, this algorithm comes equipped
with an additional set of genetic operators (RNC
mutation, Dc mutation,
Dc inversion, and Dc
IS transposition) especially developed for handling random
numerical constants (if you are not familiar with these operators,
please use the default values by clicking the Defaults button for
they work very well in all cases).
And last but not least since these parameters are crucial if you are
handling numerical constants directly, you must also choose and
adjust the range and type of numerical constants that will be used
by the GEP-RNC
algorithm during the learning process. As for the Number of
Constants per Gene parameter, a good rule of thumb consists of using
a small set of 10 different constants per gene as this seems to
provide enough diversity for most problems without inflating the
structural complexity much.
|