Predicting unknown behavior efficiently is of course the foremost goal in modeling. But extracting knowledge from the blindly designed models is becoming more and more crucial as this knowledge can be used not only to enlighten further the modeling process but also to understand the complex relationships between variables.
So, the evolutionary strategies we recommend in the APS templates
for Function Finding reflect these two main concerns: efficiency and simplicity. Basically, we recommend starting the modeling process with the
simplest learning algorithm and a simple function set well adjusted to the complexity of the problem.
APS 3.0 chooses the appropriate template for your problem according
to the number of variables in your data. This kind of template is a good starting
point that allows you to start the modeling process immediately with just a mouse click. Indeed, even if you are not familiar with evolutionary computation in general and
gene expression programming in particular, you will be able to design complex nonlinear models immediately thanks to the templates of APS 3.0. In the provided templates, all the adjustable parameters of the learning algorithm are already set and, for instance, you donít have to know how to create genetic diversity, how to set the appropriate population size, the chromosome architecture, the fitness function,
how to increase the complexity of your models, and so forth. Then, as you learn more about
APS, you will be able to explore all its modeling tools and create quickly and efficiently very good models that will allow you to understand your data like never before.
There is, however, a very important setting in APS that is not controlled by APS 3.0 templates and must be wisely chosen by you: the
number of training samples. Theoretically, if your data is well balanced and in good condition, evolutionarily
speaking the more samples the better. But there's a catch, obviously: the larger the training set the slower evolution or, in other words, the more time will be needed for generations to go by. So, you must compromise here and choose a training set with the appropriate size. A good rule of thumb consists of choosing 8-10 training samples for each independent variable in your data; all the remaining samples could be used for testing the generalizing capability of the evolved models.
So, after creating a new run you just have to click the Evolve button in the
Run Panel in order to design a model. Then you observe carefully the evolutionary process, especially the plot for the best fitness. Then, whenever you see fit, you can stop the run without fear of stopping the evolutionary process prematurely as APS 3.0 allows you to continue the evolutionary process at a later time by using the best-of-run model as the starting
point (evolve with seed). For that you just have to click on the Optimize button in the
Run Panel.
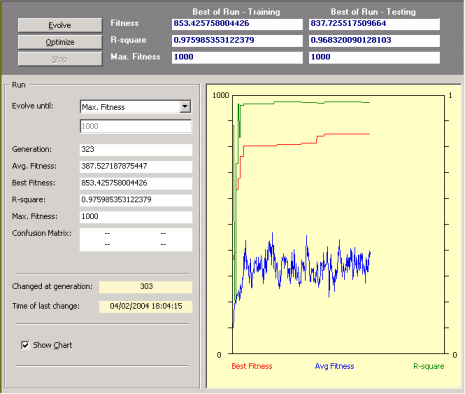
This strategy has enormous advantages as you might choose to stop the run at any time and then take a closer look at the evolved model. For instance, you can analyze its mathematical representation, its performance in the testing set,
evaluate a wide set of statistical functions for a quick and rigorous assessment of its accuracy, and so on. Then you might choose to adjust a few parameters, say, choose a different fitness function, expand the function set, add a neutral gene, and so forth, and then explore this new evolutionary path. You can repeat this process for as long as you want or until you are completely satisfied with the evolved model.
In addition, if you wish for APS 3.0 to increase the complexity of
your models automatically, you just have to
activate the Complexity Increase
Engine
and then click the Evolve button and APS will evolve better and better models composed of an increasing number of terms until no increase in best fitness takes place for a certain period of time.
|