Introduction
With in-place processing
GeneXproServer 5.0 introduces a new mode of operation that allows you to process gep files without creating job files. It is possible to process a single file or all the files under a folder as well as under any other folder there contained. In this latter case the gep files are processed in parallel. You can
Start, Continue, Simplify or Complexify the runs. In addition you can test them in several ways and also make predictions.
Even more flexibility can be attained by adding job files. When GeneXproServer finds a run it looks for a file with the same name but with the extension xml. When this file exists GeneXproServer reverts to job definition processing for that run file.
Process One File
To run, test or make predictions with a single file you open a command line and navigate to the folder where the run is. Assuming the run is called CreditApproval.gep write the following command followed by pressing enter:
gxps50c -s CreditApproval.gep 0 50
The run will start and will show an output similar to the picture below:
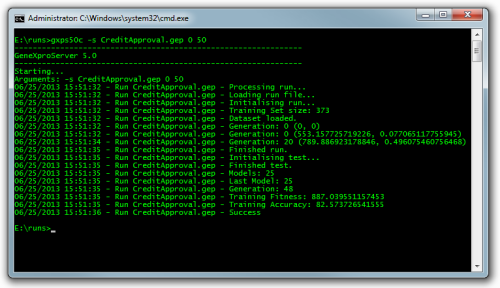
The command arguments are:
Gxps50c.exe -s [path to gep file] [Run Type (0-3)] [timeout in generations]
The description of the arguments is as follows:
-s: Process a run file.
Run Type: Whether to start a run, continue, simplify or complexify the selected model.
Values:
- Start = 0
- Continue = 1
- Simplify = 2
- Complexify = 3
Timeout: Maximum number of generations (positive integer) to run. This value is optional and should only be set if the Stop Condition in the run file is not set to Generation Number.
Example:
Continue a run using the file CreditApproval.gep and stop at 1000 generations if there isn't a lower number defined in the run.
gxps50c.exe -s c:\CreditApproval.gep 1 1000
Process All Files in Folder Tree
GeneXproServer 5.0 can process any number of runs that are in a folder as well as runs that are inside folders in that folder recursively. This is useful when you have a number of runs organized in a tree structure of folders and you need to perform one or more operations on all runs such as creating new predictions or further process the existing models.
The simple way to process runs in a folder is to use the –r command as in this example:
gxps50c -r c:\folder\ 2 1000
The example will traverse all the folders under the folder c:\folder and whenever it finds a run it will Simplify the selected model for as many generations as are defined in the run, or 1000 generations (timeout) if the run’s stop condition is not set to Generation Number.
The command arguments are:
gxps50c -r [path to folder] [run type] [timeout in generations]
The description of the arguments is as follows:
-r: Process all runs in folder recursively.
Run Type: Whether to start the runs, continue, simplify or complexify the selected model in each
run.
Values:
- Start = 0
- Continue = 1
- Simplify = 2
- Complexify = 3
Timeout: Maximum number of generations (positive integer) for each. This value is optional and should be set if the Stop Condition in the
gep files is not set to Generation Number.
Example:
gxps50c.exe -r c:\MyFolder\ 2 1000
Test One File
To test all the models of a single run you open a command line, navigate to the folder of the run and enter the following commands:
gxps50c.exe -t all both c:\MyRun.gep
This example tests all the models in the file MyRun.gep both in the training and in the validation
datasets.
The command arguments are:
gxps50c.exe -t [how many models to test] [dataset] [path to file]
The description of the arguments is as follows:
-t: Test models.
The second argument can have the following values:
all: Test all models in the run.
current: Test only the current or active model.
[number of models] : When this argument is a positive integer GeneXproServer tests that number of models starting from the last model backwards.
r[number of models] : In this format (r5 or r10, for example) GeneXproServer tests
x number of models per history in the run. Say, for example, that a run has 500 models distributed by 10 histories and this argument is set to r15, then this would direct GeneXproServer to test the last 15 models of each history in the run.
dataset: Which dataset to test.
Examples:
gxps50c.exe -t all training c:\MyRun.gep
gxps50c.exe -t current both c:\MyRun.gep
gxps50c.exe -t 10 validation c:\MyRun.gep
gxps50c.exe -t r5 validation c:\MyRun.gep
Test All Files in Folder Tree
Testing all gep files in a folder including any subfolder is very similar to the
previous point. A possible example would be:
gxps50c.exe -tr all validation c:\MyFolder\
In this example GeneXproServer will test the validation set for all the models in any gep file it finds under the MyFolder folder.
The command arguments are:
gxps50c.exe -tr [how many models to test] [dataset] [path to folder]
The description of the arguments is as follows:
-tr: Test all runs recursively.
The second argument can have the following values:
all: Test all models in the run.
current: Test only the current or active model in each run.
[number of models] : When this argument is a positive integer GeneXproServer tests that number of models starting from the last model backwards in each run.
r[number of models] : In this format (r5 or r10, for example) GeneXproServer tests
x number of models per history in the run. Say, for example, that a run has 500 models distributed by 10 histories and this argument is set to r15, then this would direct GeneXproServer to test the last 15 models of each history in the run.
dataset: Which dataset to test in each run.
Examples:
Test on both datasets all the models in each gep file in the folder c:\files\ plus any other gep
files in folders recursively:
gxps50c.exe -tr all both c:\files\
Test on the training set the current model in each gep file in the folder c:\files\ plus any other gep
files in folders recursively:
gxps50c.exe -tr current training c:\files
Test on the validation set the last 10 models in each gep file in the folder c:\files\ plus any other gep
files in folders recursively:
gxps50c.exe -tr 10 validation c:\files\
Test on the validation set the last 5 models of each run in each gep file in the folder c:\files\ plus any other gep
files in folders recursively:
gxps50c.exe -tr r5 validation c:\files\
Process and Predict One File
It is possible to process a run followed by creating predictions for a single run without using a job file. An example of that operation would be:
gxps50c.exe –spx:5 c:\TimeSeriesRun.gep 0, 1000
In this example the run TimeSeriesRun.gep will start a run from scratch with a timeout of 1000 generations (which is only applied if the stop condition of the run is not set to Generation Number) and at the end it will
make 5 predictions with the last model and save them in an xml file.
The command arguments are:
gxps50c.exe –sp[t/j/x]:[number of predictions] [path to gep file] [Run Type (0-3)]
[timeout in generations]
The description of the arguments is as follows:
-spt:n : Process run, generate
n predictions and save them in a text file.
-spj:n : Process run, generate
n predictions and save them as json in a text
file.
-spx:n : Process run, generate
n predictions and save them in an xml file.
Run Type: Whether to start a run or
continue optimizing, simplifying or complexifying the
selected model.
Values:
- Start = 0
- Continue = 1
- Simplify = 2
- Complexify = 3
Timeout: Maximum number of generations (positive integer) for each
run. This value is optional and should be set if the stop
condition in the gep file is not set to Generation Number.
Examples:
Start new run, stop at 1000 generations if there isn't a lower number defined in the run, generate
5 predictions and save them in a text file:
gxps50c.exe –spt:5 c:\TimeSeriesRun.gep 0, 1000
Improve the select model for 1000 generations if there isn't a lower number defined in the run, generate
7 predictions and save them as json in a text file:
gxps50c.exe –spj:7 c:\TimeSeriesRun.gep 1, 1000
Simplify the select model for 1000 generations if there isn't a lower number defined in the run, generate
10 predictions and save them in an xml file:
gxps50c.exe –spx:10 c:\TimeSeriesRun.gep 2, 1000
Process and Predict All Files in Folder Tree
The following example illustrates how to make predictions and process all runs in a folder (as well as its subfolders) followed by creating predictions for all runs:
gxps50c.exe –rspj:2 c:\MyFiles\ 0, 1000
In this example all the runs in the folder MyFiles
will be run from scratch with a timeout of 1000 generations (which is only applied if the stop condition of the runs is not set to Generation Number) and at the end GeneXproServer
makes 2 predictions with the last model of each run and saves them in json format in a text file. The prediction files are named after the run name with the postfix _predictions.json,
for example, DowJones_predictions.json.
The command arguments are:
gxps50c.exe –rsp[t/j/x]:[number of predictions] [path to folder] [Run Type (0-3)]
[timeout in generations]
The description of the arguments is as follows:
-rspt:n : Process all runs, generate n predictions and save them in a text file.
-rspj:n : Process all runs, generate n predictions and save them as json in a text file.
-rspx:n : Process all runs, generate n predictions and save them in an xml file.
Run Type: Whether to start a run or
continue optimizing, simplifying or complexifying the
selected model.
Values:
- Start = 0
- Continue = 1
- Simplify = 2
- Complexify = 3
Timeout: Maximum number of generations (positive integer) for each
run. This value is optional and should be set if the stop
condition in the gep files is not set to Generation Number.
Examples:
Start new runs, stop at 1000 generations if there isn't a lower number defined in the run, generate
5 predictions and save them in text files:
gxps50c.exe –rspt:5 c:\FolderWithRuns\ 0, 1000
Improve the select models for 1000 generations if there isn't a lower number defined in the runs, generate
7 predictions and save them as json in a text files:
gxps50c.exe –rspj:7 c:\ FolderWithRuns\ 1, 1000
Simplify the select models for 1000 generations if there isn't a lower number defined in the runs, generate
10 predictions and save them on xml files:
gxps50c.exe –rspx:10 c:\ FolderWithRuns\ 2, 1000
Predict One File
To make predictions using the active model in an existing run without using a job file you use the command -p. An example of that operation would be:
gxps50c.exe –pt:2 c:\TimeSeriesRun.gep
In this example the run TimeSeriesRun.gep will be loaded by GeneXproServer which will create 2 predictions using the active model and save them in a text file. The prediction files are named after the run name with the postfix _predictions,
for example, TimeSeriesRun_predictions.txt.
The command arguments are:
gxps50c.exe –p[t/j/x]:[number of predictions] [path to gep file]
The description of the arguments is as follows:
-pt:n : Generate n predictions and save them in a text file.
-pj:n : Generate n predictions and save them as json in a text file.
-px:n : Generate n predictions and save them in an xml file.
Examples:
gxps50c.exe –pt:2 c:\TimeSeriesRun.gep
gxps50c.exe –pj:3 c:\TimeSeriesRun.gep
gxps50c.exe –px:10 c:\TimeSeriesRun.gep
Predict All Files in Folder Tree
The following example illustrates how to make predictions for all runs in a folder (as well as its subfolders):
gxps50c.exe –rpj:2 c:\data\
In this example GeneXproServer makes 2 predictions with the last model of each run and saves them in json format in a text file. The prediction files are named after the run name with the postfix _predictions,
for example, DowJones_predictions.json.
The command arguments are:
gxps50c.exe –rp[t/j/x]:[number of predictions] [path to folder]
The description of the arguments is as follows:
-rpt:n : Generate n predictions and save them in a text file.
-rpj:n : Generate n predictions and save them as json in a text file.
-rpx:n : Generate n predictions and save them in an xml file.
Examples:
gxps50c.exe –rpt:2 c:\RunFolder\
gxps50c.exe –rpj:3 c:\RunFolder\
gxps50c.exe –rpx:10 c:\RunFolder\
See Also:
Related Tutorials:
Related Videos:
|

