| GeneXproTools 4.0 ships with four different Sample
Runs for Classification: Iris
Virginica, Breast Cancer, Credit
Screening, and DNA Microarrays. To try any one of them, you just
have to click its link on the Welcome Screen of GeneXproTools.
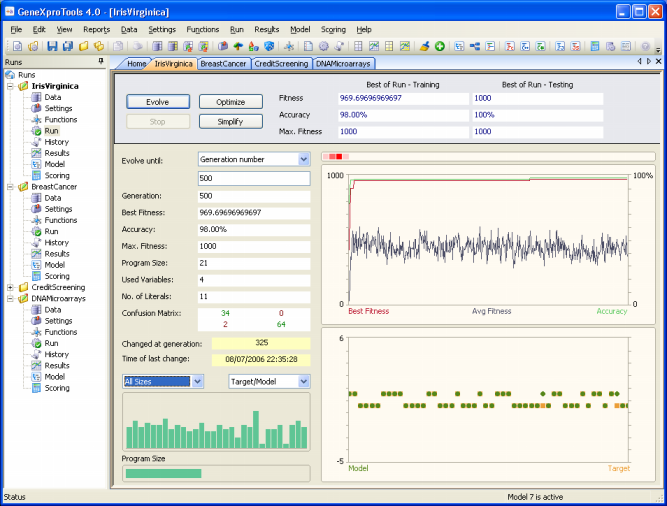
The Iris Virginica sample run is a simple real-world problem for distinguishing Iris Virginica from other two irises: Iris Setosa and Iris Versicolor. The original iris dataset contains fifty examples each of the three types of iris. In this sample run, though, the sub-problem Virginica versus Not Virginica is analyzed, where 100 randomly chosen samples are used for training and the remaining 50 for testing. You will see that this is an easy problem for GeneXproTools and exceptionally good models with 100% accuracy on the testing set can be easily created.
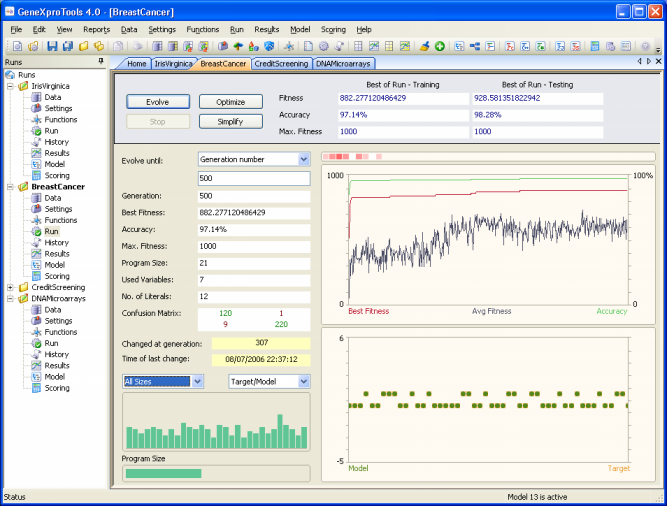
The Breast Cancer sample run is a complex real-world problem for diagnosing breast cancer based on nine different cell analysis (clump thickness, uniformity of cell size, uniformity of cell shape, marginal adhesion, single epithelial cell size, bare nuclei, bland chromatin, normal nucleoli, and mitoses). The dataset cancer1 used in this sample run was obtained from PROBEN1, with 350 samples used for training and 174 for testing. And again you will see that GeneXproTools is able to create exceptionally good models for diagnosing breast cancer with predictive accuracies around 99%.
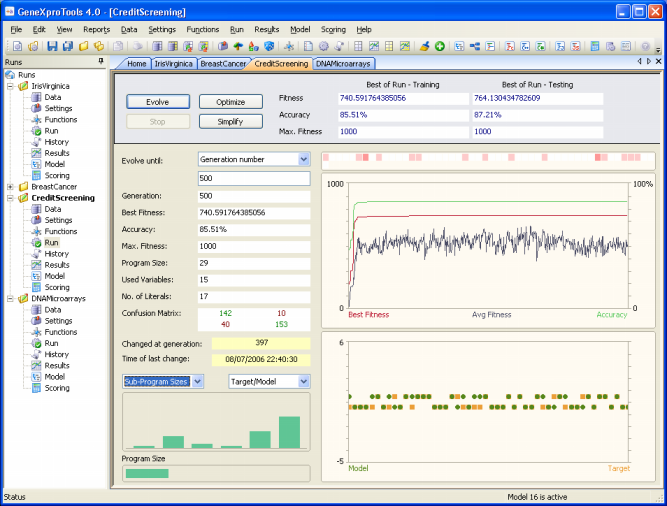
The Credit Screening sample run is a high-dimensional real-world problem. And the goal here is to decide whether to approve or not a customers request for a credit card. Each sample in the dataset represents a real credit card application and the output describes whether the bank granted the credit card or not. This problem has 51 input attributes, all of them unexplained in the original dataset for confidentiality reasons. The dataset card1 used in this sample run was obtained from PROBEN1, with 345 samples used for training and 172 for testing. And here you will see that GeneXproTools is able to create very good models with high predictive accuracy.
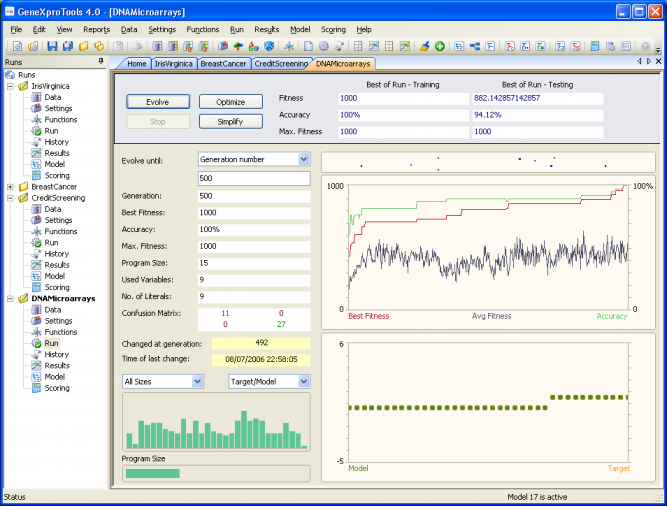
The DNA Microarrays sample run is an extremely complex real-world
problem with thousands of variables. The training dataset consists of 38 bone marrow samples (27 ALL and 11 AML), over 7129 probes from 6817 human genes.
And the testing dataset consists of 34 samples, with 20 ALL and 14 AML.
In this sample run, "0" was used to represent "ALL" and "1" to
represent "AML". The 7129 genes were numbered d0-d7128.
|