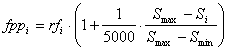| GeneXproTools 4.0 implements the Sensitivity/Specificity
fitness function both with and
without parsimony pressure. The
version with parsimony
pressure puts a little pressure on the size of the evolving
solutions, allowing the discovery of more compact models.
The Sensitivity/Specificity fitness function of GeneXproTools 4.0
is, as expected, based both on the sensitivity and
specificity.
where SEi is the sensitivity and SPi is the specificity of the individual program i, and are given by the formulas:
where TPi, TNi, FPi, and
FNi represent, respectively, the number of true
positives, true negatives, false positives, and false
negatives.
Thus, for evaluating the fitness fi of an individual program i, the following equation is used:
which obviously ranges from 0 to 1000, with 1000 corresponding to the ideal.
where Si is the size of the program, Smax and Smin represent, respectively, maximum and minimum program sizes and are evaluated by the formulas: Smax = G (h + t) Smin = G where G is the number of genes, and h and t are the head and tail sizes (note that, for simplicity, the linking function was not taken into account). Thus, when rfi = rfmax and Si = Smin (highly improbable, though, as this can only happen for very simple functions as this means that all the sub-ETs are composed of just one node), fppi = fppmax, with fppmax evaluated by the formula:
|