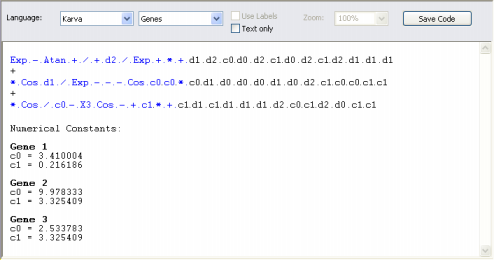
| Sometimes it is important to introduce small (or not so small) changes in an existing model, say, adjust a coefficient or
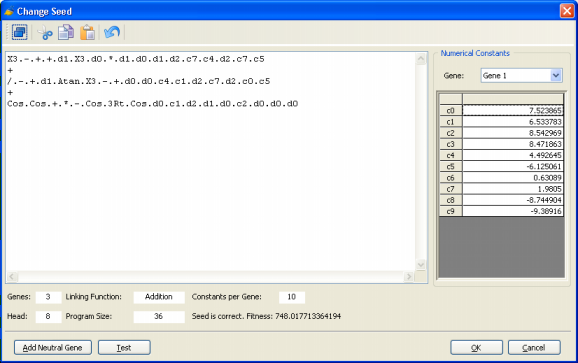
remove a term. For that you must know how to change your seed correctly. When your seed is already in Karva notation and the change is a small one, most of the times you will be able to change your seed directly in the Change Seed window and, by checking its representation as an expression tree in the Model Panel, you will be able to twinkle with it until it meets your standards.
But if the change is a big one or if your seed is not in Karva notation, then you will need to translate your model into Karva as the Change Seed window can only accept models in Karva code.
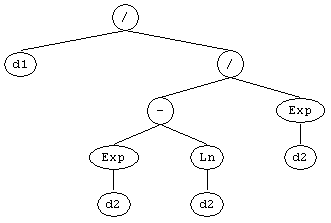
The easiest way of translating your model into Karva
code, consists of representing your model in a tree structure and then create a linear representation of the parse tree by
writing down all the nodes in the tree from top to bottom and from left to right. You will end up with a
K-expression which you can, with only a small modification, use as seed.
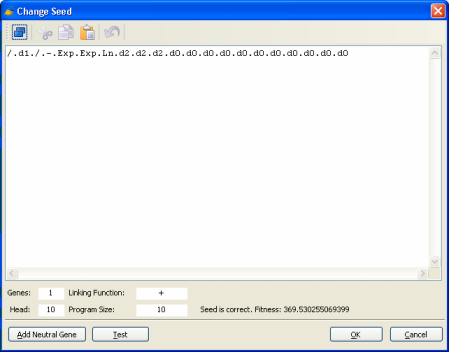
can be represented linearly by the following K-expression: /.d1./.-.Exp.Exp.Ln.d2.d2.d2 Now you only need to incorporate this K-expression into a correct gene structure, with a head and a tail correctly defined. A simple way of doing this consists of considering the entire K-expression the head of your gene and then add a tail (with the correct length, of course) containing only terminals (which terminals you add is irrelevant as these terminals will not interfere with the expression of your K-expression). For instance, the K-expression above could be correctly accommodated into the following gene (the head is shown in blue): /.d1./.-.Exp.Exp.Ln.d2.d2.d2.d0.d0.d0.d0.d0.d0.d0.d0.d0.d0.d0 Then you just have to create a new run in GeneXproTools and evolve quickly (for only one generation is enough) a proto-model with exactly the same structure you’ll need for integrating your seed model (that is, number of genes, head size, linking function, function set, etc.), and then replace this proto-model in the Change Seed window by your seed.
These are only some examples of the things you can accomplish with the Change Seed method of
GeneXproTools 4.0. If you want to make other kinds of modification,
for instance, use different
K-expressions to represent different terms, then you must be familiar with the architectures of
GeneXproTools 4.0 learning algorithms so that you can map correctly all your modifications into
Karva notation. |