|
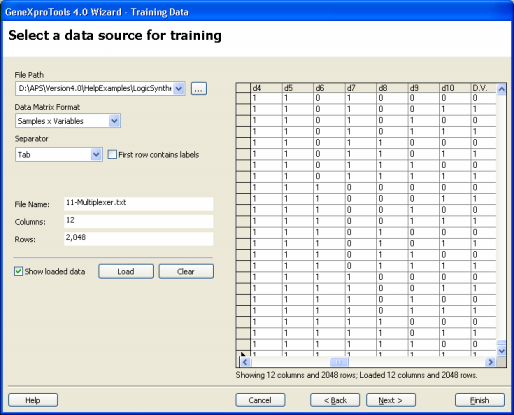
Before designing a circuit with GeneXproTools 4.0 you must first load the
truth tables for the learning algorithm. The data screening engine of GeneXproTools 4.0 checks the validity of all the
truth tables and is operative every time you load data sets
either for training or testing. Only the values of False and True
(case insensitive) or "0" and "1" are valid as
input. Note,
however, that GeneXproTools uses 0's and 1's internally and,
therefore, False and True are respectively converted into
"0" and "1" and shown in this format. Missing or
invalid values can be detected by the data screening engine so that
you can identify these faulty samples and correct them. GeneXproTools 4.0 allows you to work either with databases/Excel or text files and, for text files, accepts two different data matrix formats. The first is the standard Samples x Variables format where samples are in rows and variables in columns, with the dependent variable or function output occupying the rightmost position. In the small example below with eight samples, OUTPUT is the function output and A, B, and C are the independent variables: A B C OUTPUT 0 0 0 0 0 0 1 0 0 1 0 0 0 1 1 1 1 0 0 0 1 0 1 1 1 1 0 1 1 1 1 1 And the second, is the Gene Expression Matrix format commonly used in DNA microarrays studies where samples are in columns and variables in rows, with the function output occupying the topmost position. For instance, in Gene Expression Matrix format, the truth table above corresponds to: OUTPUT 0 0 0 1 0 1 1 1 A 0 0 0 0 1 1 1 1 B 0 0 1 1 0 0 1 1 C 0 1 0 1 0 1 0 1 which is very handy for datasets with a relatively small number of samples and thousands of variables. Note, however, that for Excel files this format is not supported and if your data is kept in this format in Excel, you must copy it to a text file so that it can be loaded into GeneXproTools. GeneXproTools uses the Samples x Variables format throughout and therefore all formats are automatically converted and shown in this format. GeneXproTools supports the standard separators (space, tab, comma, semicolon, and pipe) and detects them automatically. The use of labels to identify your variables is optional and GeneXproTools also detects automatically whether they are present or not. If you use them, however, you will be able to generate more intelligible code where each variable is identified by its name, by checking the Use Labels box in the Model Panel.
To Load Truth Tables for Modeling
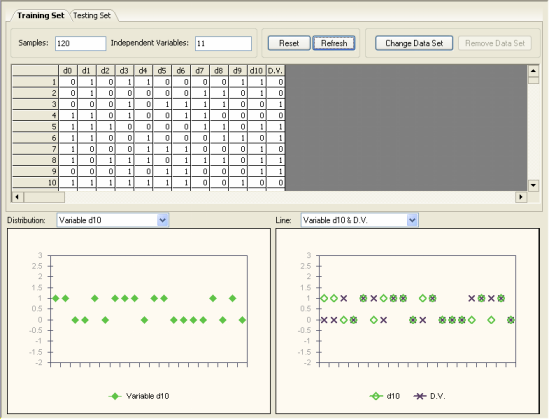
After loading your data into GeneXproTools, in the Data Panel you can visualize the distribution of values for each variable and also plot each independent variable against the function output or dependent variable.
|