| Except for the different types of linking functions,
the General Settings Tabs used both for Function Finding, Classification, and
Logic Synthesis are exactly the same and look like this:
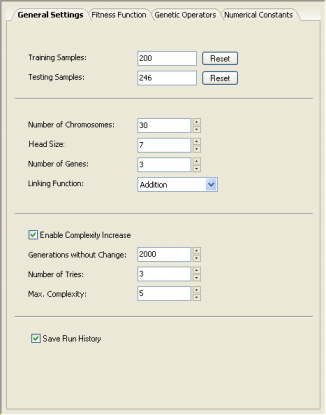
Here you can change the number of samples used for training and the
number of samples used for testing the generalizing capabilities of
the evolved models.
Here is also the place where you choose the size of the population
of evolving models (Number of Chromosomes) and where you design the structural organization
of the chromosomes by choosing the
Head Size, the Number of Genes, and the Linking Function. For
mathematical problems, you can choose from four different linking
functions (addition, subtraction, multiplication, and division); and
for Boolean problems, you can choose from 10 different linking
functions (And, Or, Nand, Nor, Xor, Nxor, Less Than, Greater Than,
Less Or Equal, Greater Or Equal).
The
Head Size and the Number of Genes are constrained by the maximum
chromosome size allowed in GeneXproTools, which is 2049. And the chromosome
size depends not only on the Number of Genes and Head Size but also
on maximum arity and the learning
algorithm (with or without random numerical constants).
And, finally, in the General Settings Tab
you can also enter the parameters for the Complexity Increase
Engine. In the Generations Without Change box you set the period of time you think acceptable for evolution to occur without improvement in best fitness, after which
a mass extinction or a neutral gene (an extra term) is automatically added to your model;
in the Number of Tries box you enter the number of consecutive evolutionary epochs (defined by the
parameter Generations Without Change) you will allow before a
neutral gene is introduced in all evolving models; in the Max. Complexity box you write the maximum number of terms (genes) you’ll allow in your model and no other terms will be introduced beyond this threshold.
The Complexity Increase Engine of GeneXproTools 4.0 might be a very powerful modeling tool, especially for Time Series Prediction where good models could take longer to come by, but you must be careful not to create excessively complex models for, most of the times, a greater complexity does not
necessarily imply a greater efficiency.
The General Settings Tab used for Time Series Prediction gives access to all the parameters listed above, but has also additional parameters necessary for dealing with time series data
(the Embedding Dimension, the Delay Time, the Prediction Mode and the number of
Testing Predictions).
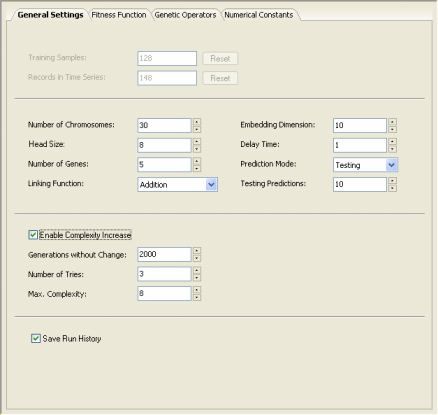
In practice, the embedding dimension corresponds to the number of
independent variables (terminals) after your time series has been
transformed. The delay time t
determines how data are processed, that is, continuously if t
= 1 or at t intervals.
These two parameters, together with the size of the time series and
the prediction mode, will determine the final number of training
samples after the transformation of the time series.
|

